Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
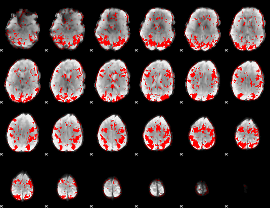 |
GLM binary result thresholded by 2.5
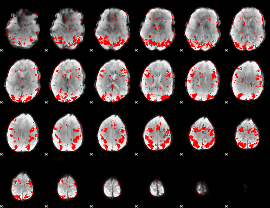 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
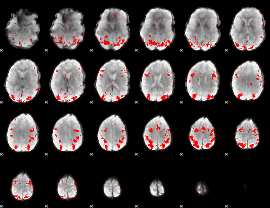 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
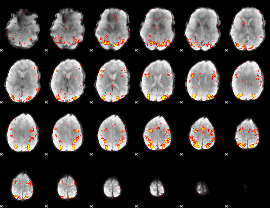 |
GLM Z-score result thresholded by 4
 |
Component:123; Jaccard:0.11373
 |
Component:123; Jaccard:0.13956
 |
Component:123; Jaccard:0.17222
 |
Component:123; Jaccard:0.20749
 |
Component:123; Jaccard:0.25003
 |
Component:123; Jaccard:0.28694
 |
Component:123; Jaccard:0.30946
 |
Correlation:0.30015
  |
Correlation:0.30015
  |
Correlation:0.30015
  |
Correlation:0.30015
  |
Correlation:0.30015
  |
Correlation:0.30015
  |
Correlation:0.30015
  |
Component:276; Jaccard:0.097674
 |
Component:049; Jaccard:0.10863
 |
Component:049; Jaccard:0.13426
 |
Component:049; Jaccard:0.16475
 |
Component:049; Jaccard:0.19942
 |
Component:049; Jaccard:0.23357
 |
Component:049; Jaccard:0.25866
 |
Correlation:0.29098
  |
Correlation:0.28015
  |
Correlation:0.28015
  |
Correlation:0.28015
  |
Correlation:0.28015
  |
Correlation:0.28015
  |
Correlation:0.28015
  |
Component:330; Jaccard:0.090281
 |
Component:276; Jaccard:0.10833
 |
Component:276; Jaccard:0.1189
 |
Component:276; Jaccard:0.12928
 |
Component:276; Jaccard:0.13772
 |
Component:327; Jaccard:0.14423
 |
Component:378; Jaccard:0.15041
 |
Correlation:0.38518
  |
Correlation:0.29098
  |
Correlation:0.29098
  |
Correlation:0.29098
  |
Correlation:0.29098
  |
Correlation:0.03208
  |
Correlation:0.24328
  |
Component:111; Jaccard:0.089594
 |
Component:330; Jaccard:0.099017
 |
Component:327; Jaccard:0.11123
 |
Component:327; Jaccard:0.12506
 |
Component:327; Jaccard:0.13702
 |
Component:378; Jaccard:0.14179
 |
Component:327; Jaccard:0.14662
 |
Correlation:0.2162
  |
Correlation:0.38518
  |
Correlation:0.03208
  |
Correlation:0.03208
  |
Correlation:0.03208
  |
Correlation:0.24328
  |
Correlation:0.03208
  |
Component:049; Jaccard:0.088217
 |
Component:327; Jaccard:0.098368
 |
Component:330; Jaccard:0.10754
 |
Component:378; Jaccard:0.11577
 |
Component:378; Jaccard:0.12873
 |
Component:276; Jaccard:0.14137
 |
Component:276; Jaccard:0.14052
 |
Correlation:0.28015
  |
Correlation:0.03208
  |
Correlation:0.38518
  |
Correlation:0.24328
  |
Correlation:0.24328
  |
Correlation:0.29098
  |
Correlation:0.29098
  |
Component:327; Jaccard:0.086137
 |
Component:111; Jaccard:0.095639
 |
Component:378; Jaccard:0.10346
 |
Component:330; Jaccard:0.11499
 |
Component:330; Jaccard:0.1209
 |
Component:330; Jaccard:0.12409
 |
Component:330; Jaccard:0.12023
 |
Correlation:0.03208
  |
Correlation:0.2162
  |
Correlation:0.24328
  |
Correlation:0.38518
  |
Correlation:0.38518
  |
Correlation:0.38518
  |
Correlation:0.38518
  |
Component:274; Jaccard:0.08505
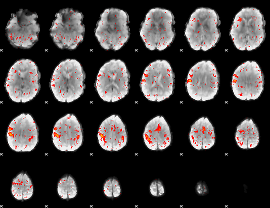 |
Component:274; Jaccard:0.092026
 |
Component:111; Jaccard:0.10118
 |
Component:111; Jaccard:0.10379
 |
Component:111; Jaccard:0.10407
 |
Component:274; Jaccard:0.10672
 |
Component:299; Jaccard:0.10586
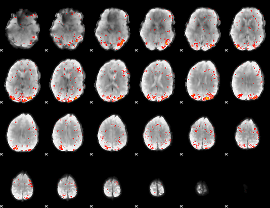 |
Correlation:0.21159
  |
Correlation:0.21159
  |
Correlation:0.2162
  |
Correlation:0.2162
  |
Correlation:0.2162
  |
Correlation:0.21159
  |
Correlation:0.20845
  |
Component:239; Jaccard:0.08485
 |
Component:378; Jaccard:0.090992
 |
Component:274; Jaccard:0.097956
 |
Component:274; Jaccard:0.10162
 |
Component:274; Jaccard:0.10406
 |
Component:299; Jaccard:0.10335
 |
Component:274; Jaccard:0.10115
 |
Correlation:0.33374
  |
Correlation:0.24328
  |
Correlation:0.21159
  |
Correlation:0.21159
  |
Correlation:0.21159
  |
Correlation:0.20845
  |
Correlation:0.21159
  |
Component:141; Jaccard:0.079677
 |
Component:239; Jaccard:0.089759
 |
Component:239; Jaccard:0.09506
 |
Component:239; Jaccard:0.099007
 |
Component:239; Jaccard:0.10117
 |
Component:239; Jaccard:0.10005
 |
Component:239; Jaccard:0.093127
 |
Correlation:0.54175
  |
Correlation:0.33374
  |
Correlation:0.33374
  |
Correlation:0.33374
  |
Correlation:0.33374
  |
Correlation:0.33374
  |
Correlation:0.33374
  |
Component:378; Jaccard:0.078924
 |
Component:055; Jaccard:0.084036
 |
Component:055; Jaccard:0.086244
 |
Component:299; Jaccard:0.092353
 |
Component:299; Jaccard:0.098465
 |
Component:111; Jaccard:0.099959
 |
Component:210; Jaccard:0.092604
 |
Correlation:0.24328
  |
Correlation:0.092002
  |
Correlation:0.092002
  |
Correlation:0.20845
  |
Correlation:0.20845
  |
Correlation:0.2162
  |
Correlation:0.074645
  |
Cope: 02:REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
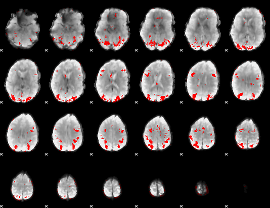
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
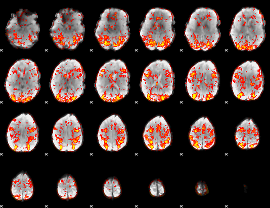
GLM Z-score result thresholded by 1
 |
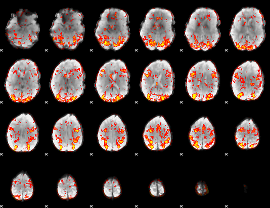
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
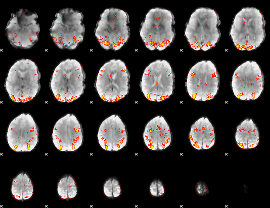
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
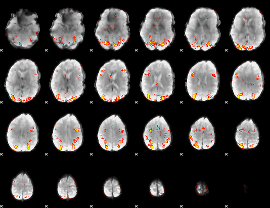 |
GLM Z-score result thresholded by 4
 |
Component:123; Jaccard:0.1345
 |
Component:123; Jaccard:0.15985
 |
Component:123; Jaccard:0.18747
 |
Component:123; Jaccard:0.21412
 |
Component:123; Jaccard:0.23202
 |
Component:123; Jaccard:0.2384
 |
Component:049; Jaccard:0.23812
 |
Correlation:-0.047876
  |
Correlation:-0.047876
  |
Correlation:-0.047876
  |
Correlation:-0.047876
  |
Correlation:-0.047876
  |
Correlation:-0.047876
  |
Correlation:0.097086
  |
Component:049; Jaccard:0.11191
 |
Component:049; Jaccard:0.13447
 |
Component:049; Jaccard:0.15981
 |
Component:049; Jaccard:0.18785
 |
Component:049; Jaccard:0.21368
 |
Component:049; Jaccard:0.23069
 |
Component:123; Jaccard:0.22866
 |
Correlation:0.097086
  |
Correlation:0.097086
  |
Correlation:0.097086
  |
Correlation:0.097086
  |
Correlation:0.097086
  |
Correlation:0.097086
  |
Correlation:-0.047876
  |
Component:378; Jaccard:0.099881
 |
Component:378; Jaccard:0.11521
 |
Component:378; Jaccard:0.13168
 |
Component:378; Jaccard:0.14699
 |
Component:378; Jaccard:0.16029
 |
Component:378; Jaccard:0.16406
 |
Component:378; Jaccard:0.16502
 |
Correlation:0.21586
  |
Correlation:0.21586
  |
Correlation:0.21586
  |
Correlation:0.21586
  |
Correlation:0.21586
  |
Correlation:0.21586
  |
Correlation:0.21586
  |
Component:330; Jaccard:0.098588
 |
Component:327; Jaccard:0.10534
 |
Component:327; Jaccard:0.11531
 |
Component:327; Jaccard:0.12515
 |
Component:327; Jaccard:0.13112
 |
Component:327; Jaccard:0.13415
 |
Component:327; Jaccard:0.13171
 |
Correlation:-0.14321
  |
Correlation:0.20303
  |
Correlation:0.20303
  |
Correlation:0.20303
  |
Correlation:0.20303
  |
Correlation:0.20303
  |
Correlation:0.20303
  |
Component:327; Jaccard:0.094771
 |
Component:330; Jaccard:0.10479
 |
Component:210; Jaccard:0.11068
 |
Component:210; Jaccard:0.11767
 |
Component:210; Jaccard:0.12045
 |
Component:210; Jaccard:0.11691
 |
Component:210; Jaccard:0.10813
 |
Correlation:0.20303
  |
Correlation:-0.14321
  |
Correlation:0.14597
  |
Correlation:0.14597
  |
Correlation:0.14597
  |
Correlation:0.14597
  |
Correlation:0.14597
  |
Component:210; Jaccard:0.091184
 |
Component:210; Jaccard:0.10116
 |
Component:330; Jaccard:0.10896
 |
Component:330; Jaccard:0.11055
 |
Component:299; Jaccard:0.10684
 |
Component:299; Jaccard:0.10716
 |
Component:299; Jaccard:0.10307
 |
Correlation:0.14597
  |
Correlation:0.14597
  |
Correlation:-0.14321
  |
Correlation:-0.14321
  |
Correlation:0.10536
  |
Correlation:0.10536
  |
Correlation:0.10536
  |
Component:276; Jaccard:0.089431
 |
Component:276; Jaccard:0.09591
 |
Component:276; Jaccard:0.1013
 |
Component:276; Jaccard:0.10461
 |
Component:330; Jaccard:0.10568
 |
Component:172; Jaccard:0.10076
 |
Component:172; Jaccard:0.092253
 |
Correlation:-0.090049
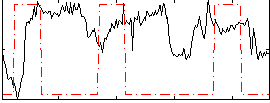  |
Correlation:-0.090049
  |
Correlation:-0.090049
  |
Correlation:-0.090049
  |
Correlation:-0.14321
  |
Correlation:0.318
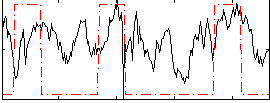  |
Correlation:0.318
  |
Component:029; Jaccard:0.08295
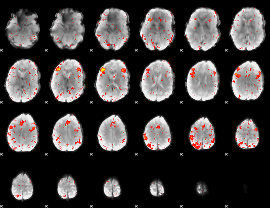 |
Component:359; Jaccard:0.091173
 |
Component:359; Jaccard:0.099558
 |
Component:359; Jaccard:0.1024
 |
Component:276; Jaccard:0.10436
 |
Component:276; Jaccard:0.098857
 |
Component:276; Jaccard:0.089845
 |
Correlation:-0.097114
  |
Correlation:0.13261
 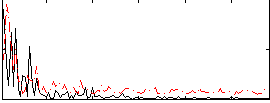 |
Correlation:0.13261
  |
Correlation:0.13261
  |
Correlation:-0.090049
  |
Correlation:-0.090049
  |
Correlation:-0.090049
  |
Component:111; Jaccard:0.082793
 |
Component:299; Jaccard:0.087567
 |
Component:299; Jaccard:0.095506
 |
Component:299; Jaccard:0.10185
 |
Component:172; Jaccard:0.1038
 |
Component:330; Jaccard:0.09632
 |
Component:330; Jaccard:0.08276
 |
Correlation:-0.10369
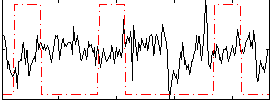  |
Correlation:0.10536
  |
Correlation:0.10536
  |
Correlation:0.10536
  |
Correlation:0.318
  |
Correlation:-0.14321
  |
Correlation:-0.14321
  |
Component:274; Jaccard:0.080988
 |
Component:172; Jaccard:0.087358
 |
Component:172; Jaccard:0.094954
 |
Component:172; Jaccard:0.10113
 |
Component:359; Jaccard:0.10247
 |
Component:359; Jaccard:0.093911
 |
Component:359; Jaccard:0.079517
 |
Correlation:-0.083335
 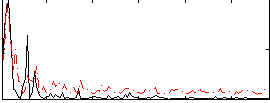 |
Correlation:0.318
  |
Correlation:0.318
  |
Correlation:0.318
  |
Correlation:0.13261
  |
Correlation:0.13261
  |
Correlation:0.13261
  |
Cope: 03:MATCH-REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
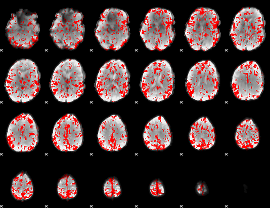 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
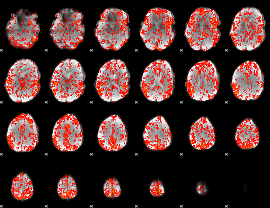 |
GLM Z-score result thresholded by 2
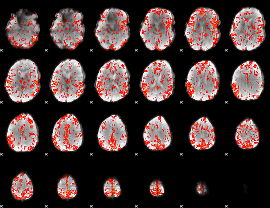 |
GLM Z-score result thresholded by 2.5
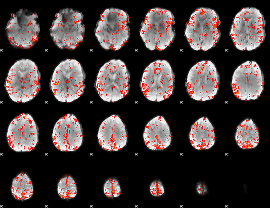 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:253; Jaccard:0.089746
 |
Component:253; Jaccard:0.11094
 |
Component:253; Jaccard:0.13603
 |
Component:253; Jaccard:0.15827
 |
Component:253; Jaccard:0.16107
 |
Component:253; Jaccard:0.13258
 |
Component:253; Jaccard:0.083
 |
Correlation:0.34288
  |
Correlation:0.34288
  |
Correlation:0.34288
  |
Correlation:0.34288
  |
Correlation:0.34288
  |
Correlation:0.34288
  |
Correlation:0.34288
  |
Component:384; Jaccard:0.08388
 |
Component:384; Jaccard:0.094388
 |
Component:207; Jaccard:0.10609
 |
Component:207; Jaccard:0.1236
 |
Component:207; Jaccard:0.12584
 |
Component:207; Jaccard:0.10738
 |
Component:207; Jaccard:0.065324
 |
Correlation:0.4142
  |
Correlation:0.4142
  |
Correlation:0.027716
  |
Correlation:0.027716
  |
Correlation:0.027716
  |
Correlation:0.027716
  |
Correlation:0.027716
  |
Component:045; Jaccard:0.081618
 |
Component:045; Jaccard:0.091397
 |
Component:156; Jaccard:0.10412
 |
Component:156; Jaccard:0.11133
 |
Component:156; Jaccard:0.10798
 |
Component:156; Jaccard:0.088146
 |
Component:156; Jaccard:0.057107
 |
Correlation:0.30256
  |
Correlation:0.30256
  |
Correlation:0.1315
 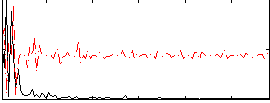 |
Correlation:0.1315
  |
Correlation:0.1315
  |
Correlation:0.1315
  |
Correlation:0.1315
  |
Component:303; Jaccard:0.081509
 |
Component:156; Jaccard:0.091194
 |
Component:384; Jaccard:0.10324
 |
Component:384; Jaccard:0.10797
 |
Component:384; Jaccard:0.10265
 |
Component:384; Jaccard:0.078879
 |
Component:384; Jaccard:0.04977
 |
Correlation:0.30323
  |
Correlation:0.1315
  |
Correlation:0.4142
  |
Correlation:0.4142
  |
Correlation:0.4142
  |
Correlation:0.4142
  |
Correlation:0.4142
  |
Component:026; Jaccard:0.078008
 |
Component:026; Jaccard:0.091128
 |
Component:026; Jaccard:0.10185
 |
Component:026; Jaccard:0.1071
 |
Component:026; Jaccard:0.098374
 |
Component:026; Jaccard:0.074323
 |
Component:026; Jaccard:0.045166
 |
Correlation:0.24979
  |
Correlation:0.24979
  |
Correlation:0.24979
  |
Correlation:0.24979
  |
Correlation:0.24979
  |
Correlation:0.24979
  |
Correlation:0.24979
  |
Component:156; Jaccard:0.077844
 |
Component:207; Jaccard:0.087478
 |
Component:045; Jaccard:0.098296
 |
Component:045; Jaccard:0.099469
 |
Component:400; Jaccard:0.090753
 |
Component:400; Jaccard:0.071525
 |
Component:400; Jaccard:0.044921
 |
Correlation:0.1315
  |
Correlation:0.027716
  |
Correlation:0.30256
  |
Correlation:0.30256
  |
Correlation:0.11089
  |
Correlation:0.11089
  |
Correlation:0.11089
  |
Component:091; Jaccard:0.077362
 |
Component:021; Jaccard:0.085624
 |
Component:021; Jaccard:0.094523
 |
Component:021; Jaccard:0.097877
 |
Component:021; Jaccard:0.089604
 |
Component:388; Jaccard:0.0675
 |
Component:010; Jaccard:0.042471
 |
Correlation:0.34295
  |
Correlation:0.39838
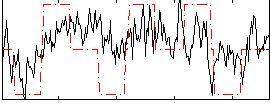  |
Correlation:0.39838
  |
Correlation:0.39838
  |
Correlation:0.39838
  |
Correlation:0.02748
  |
Correlation:0.44196
  |
Component:252; Jaccard:0.07579
 |
Component:303; Jaccard:0.083583
 |
Component:252; Jaccard:0.089427
 |
Component:400; Jaccard:0.094862
 |
Component:045; Jaccard:0.087919
 |
Component:045; Jaccard:0.066088
 |
Component:252; Jaccard:0.042432
 |
Correlation:0.22264
  |
Correlation:0.30323
  |
Correlation:0.22264
  |
Correlation:0.11089
  |
Correlation:0.30256
  |
Correlation:0.30256
  |
Correlation:0.22264
  |
Component:263; Jaccard:0.075587
 |
Component:252; Jaccard:0.0834
 |
Component:385; Jaccard:0.089323
 |
Component:388; Jaccard:0.091581
 |
Component:388; Jaccard:0.08666
 |
Component:021; Jaccard:0.065852
 |
Component:388; Jaccard:0.040694
 |
Correlation:0.50234
  |
Correlation:0.22264
  |
Correlation:0.31945
 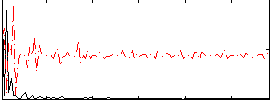 |
Correlation:0.02748
  |
Correlation:0.02748
  |
Correlation:0.39838
  |
Correlation:0.02748
  |
Component:021; Jaccard:0.074934
 |
Component:263; Jaccard:0.082168
 |
Component:400; Jaccard:0.087997
 |
Component:252; Jaccard:0.091445
 |
Component:252; Jaccard:0.081354
 |
Component:010; Jaccard:0.06432
 |
Component:385; Jaccard:0.040253
 |
Correlation:0.39838
  |
Correlation:0.50234
  |
Correlation:0.11089
  |
Correlation:0.22264
  |
Correlation:0.22264
  |
Correlation:0.44196
  |
Correlation:0.31945
  |
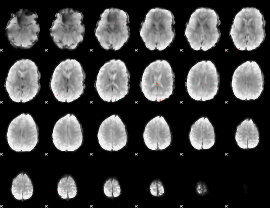
Cope: 04:REL-MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
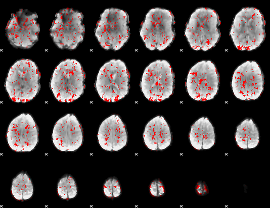 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:172; Jaccard:0.084334
 |
Component:172; Jaccard:0.081832
 |
Component:172; Jaccard:0.068054
 |
Component:172; Jaccard:0.042642
 |
Component:172; Jaccard:0.023703
 |
Component:172; Jaccard:0.012614
 |
Component:172; Jaccard:0.005487
 |
Correlation:0.27221
  |
Correlation:0.27221
  |
Correlation:0.27221
  |
Correlation:0.27221
  |
Correlation:0.27221
  |
Correlation:0.27221
  |
Correlation:0.27221
  |
Component:352; Jaccard:0.064585
 |
Component:286; Jaccard:0.058081
 |
Component:286; Jaccard:0.045178
 |
Component:352; Jaccard:0.026561
 |
Component:352; Jaccard:0.012453
 |
Component:352; Jaccard:0.006013
 |
Component:352; Jaccard:0.002246
 |
Correlation:0.12724
  |
Correlation:0.041993
  |
Correlation:0.041993
  |
Correlation:0.12724
  |
Correlation:0.12724
  |
Correlation:0.12724
  |
Correlation:0.12724
  |
Component:286; Jaccard:0.060146
 |
Component:352; Jaccard:0.057584
 |
Component:352; Jaccard:0.044625
 |
Component:286; Jaccard:0.024034
 |
Component:210; Jaccard:0.011231
 |
Component:117; Jaccard:0.004857
 |
Component:117; Jaccard:0.002013
 |
Correlation:0.041993
  |
Correlation:0.12724
  |
Correlation:0.12724
  |
Correlation:0.041993
  |
Correlation:0.042274
  |
Correlation:-0.027685
  |
Correlation:-0.027685
  |
Component:117; Jaccard:0.057801
 |
Component:117; Jaccard:0.052407
 |
Component:117; Jaccard:0.039374
 |
Component:210; Jaccard:0.023296
 |
Component:117; Jaccard:0.010456
 |
Component:210; Jaccard:0.004259
 |
Component:296; Jaccard:0.001556
 |
Correlation:-0.027685
  |
Correlation:-0.027685
  |
Correlation:-0.027685
  |
Correlation:0.042274
  |
Correlation:-0.027685
  |
Correlation:0.042274
  |
Correlation:-0.18246
  |
Component:251; Jaccard:0.055075
 |
Component:210; Jaccard:0.050853
 |
Component:210; Jaccard:0.038919
 |
Component:117; Jaccard:0.020018
 |
Component:286; Jaccard:0.008412
 |
Component:061; Jaccard:0.003042
 |
Component:103; Jaccard:0.001465
 |
Correlation:-0.084316
  |
Correlation:0.042274
  |
Correlation:0.042274
  |
Correlation:-0.027685
  |
Correlation:0.041993
  |
Correlation:-0.009832
  |
Correlation:-0.063068
  |
Component:210; Jaccard:0.054922
 |
Component:200; Jaccard:0.044668
 |
Component:082; Jaccard:0.031601
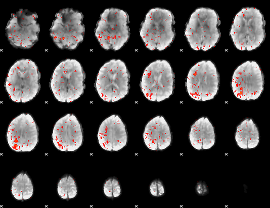 |
Component:200; Jaccard:0.016505
 |
Component:209; Jaccard:0.006919
 |
Component:103; Jaccard:0.002841
 |
Component:038; Jaccard:0.00144
 |
Correlation:0.042274
  |
Correlation:-0.061404
  |
Correlation:0.054481
  |
Correlation:-0.061404
  |
Correlation:0.014798
  |
Correlation:-0.063068
  |
Correlation:-0.083427
  |
Component:240; Jaccard:0.052308
 |
Component:082; Jaccard:0.04436
 |
Component:200; Jaccard:0.030897
 |
Component:139; Jaccard:0.016378
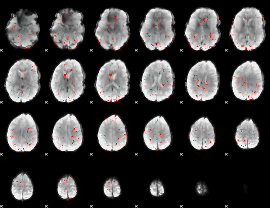 |
Component:139; Jaccard:0.006833
 |
Component:209; Jaccard:0.002735
 |
Component:267; Jaccard:0.001363
 |
Correlation:0.19275
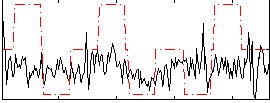  |
Correlation:0.054481
  |
Correlation:-0.061404
  |
Correlation:0.16954
  |
Correlation:0.16954
  |
Correlation:0.014798
  |
Correlation:-0.096654
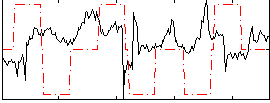 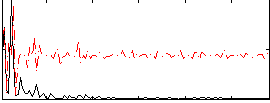 |
Component:200; Jaccard:0.051142
 |
Component:240; Jaccard:0.043195
 |
Component:240; Jaccard:0.029081
 |
Component:082; Jaccard:0.01609
 |
Component:240; Jaccard:0.006794
 |
Component:267; Jaccard:0.002696
 |
Component:333; Jaccard:0.001288
 |
Correlation:-0.061404
  |
Correlation:0.19275
  |
Correlation:0.19275
  |
Correlation:0.054481
  |
Correlation:0.19275
  |
Correlation:-0.096654
  |
Correlation:0.057655
 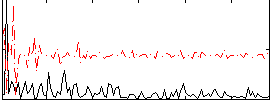 |
Component:083; Jaccard:0.050909
 |
Component:251; Jaccard:0.042474
 |
Component:225; Jaccard:0.029055
 |
Component:240; Jaccard:0.014544
 |
Component:200; Jaccard:0.006682
 |
Component:193; Jaccard:0.002513
 |
Component:269; Jaccard:0.001287
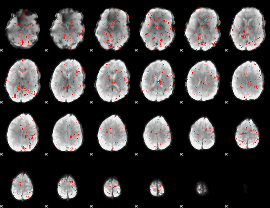 |
Correlation:-0.18222
  |
Correlation:-0.084316
  |
Correlation:0.30828
  |
Correlation:0.19275
  |
Correlation:-0.061404
  |
Correlation:-0.33986
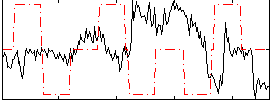  |
Correlation:0.025342
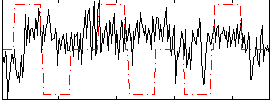  |
Component:082; Jaccard:0.049608
 |
Component:359; Jaccard:0.041592
 |
Component:359; Jaccard:0.02797
 |
Component:359; Jaccard:0.014154
 |
Component:061; Jaccard:0.006585
 |
Component:200; Jaccard:0.002475
 |
Component:280; Jaccard:0.001249
 |
Correlation:0.054481
  |
Correlation:0.16213
  |
Correlation:0.16213
  |
Correlation:0.16213
  |
Correlation:-0.009832
  |
Correlation:-0.061404
  |
Correlation:-0.0895
  |
Cope: 05:neg_MATCH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
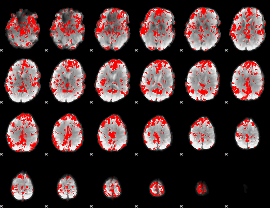 |
GLM binary result thresholded by 2.5
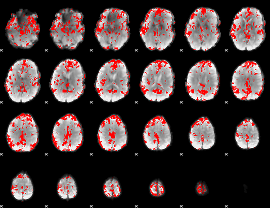 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
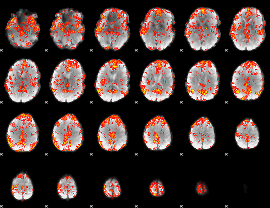 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
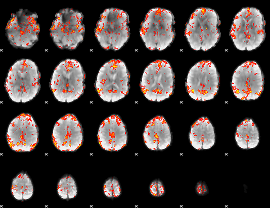 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:328; Jaccard:0.11659
 |
Component:328; Jaccard:0.13649
 |
Component:328; Jaccard:0.15735
 |
Component:328; Jaccard:0.17731
 |
Component:350; Jaccard:0.1932
 |
Component:350; Jaccard:0.20994
 |
Component:350; Jaccard:0.21293
 |
Correlation:0.058791
  |
Correlation:0.058791
  |
Correlation:0.058791
  |
Correlation:0.058791
  |
Correlation:-0.10214
  |
Correlation:-0.10214
  |
Correlation:-0.10214
  |
Component:138; Jaccard:0.11213
 |
Component:350; Jaccard:0.12774
 |
Component:350; Jaccard:0.14941
 |
Component:350; Jaccard:0.17065
 |
Component:328; Jaccard:0.19268
 |
Component:328; Jaccard:0.1958
 |
Component:328; Jaccard:0.18987
 |
Correlation:-0.078284
  |
Correlation:-0.10214
  |
Correlation:-0.10214
  |
Correlation:-0.10214
  |
Correlation:0.058791
  |
Correlation:0.058791
  |
Correlation:0.058791
  |
Component:168; Jaccard:0.11099
 |
Component:138; Jaccard:0.12737
 |
Component:138; Jaccard:0.14233
 |
Component:138; Jaccard:0.15391
 |
Component:320; Jaccard:0.16955
 |
Component:320; Jaccard:0.17693
 |
Component:320; Jaccard:0.17196
 |
Correlation:0.15369
  |
Correlation:-0.078284
  |
Correlation:-0.078284
  |
Correlation:-0.078284
  |
Correlation:0.14535
  |
Correlation:0.14535
  |
Correlation:0.14535
  |
Component:350; Jaccard:0.10825
 |
Component:168; Jaccard:0.1253
 |
Component:168; Jaccard:0.13906
 |
Component:320; Jaccard:0.15345
 |
Component:168; Jaccard:0.159
 |
Component:168; Jaccard:0.16009
 |
Component:168; Jaccard:0.1515
 |
Correlation:-0.10214
  |
Correlation:0.15369
  |
Correlation:0.15369
  |
Correlation:0.14535
  |
Correlation:0.15369
  |
Correlation:0.15369
  |
Correlation:0.15369
  |
Component:263; Jaccard:0.10587
 |
Component:242; Jaccard:0.11957
 |
Component:320; Jaccard:0.13579
 |
Component:168; Jaccard:0.15123
 |
Component:138; Jaccard:0.15657
 |
Component:138; Jaccard:0.15093
 |
Component:361; Jaccard:0.14505
 |
Correlation:-0.20267
  |
Correlation:0.030015
  |
Correlation:0.14535
  |
Correlation:0.15369
  |
Correlation:-0.078284
  |
Correlation:-0.078284
  |
Correlation:-0.003947
  |
Component:242; Jaccard:0.10493
 |
Component:320; Jaccard:0.11698
 |
Component:242; Jaccard:0.13318
 |
Component:242; Jaccard:0.14452
 |
Component:242; Jaccard:0.15081
 |
Component:361; Jaccard:0.14809
 |
Component:138; Jaccard:0.13738
 |
Correlation:0.030015
  |
Correlation:0.14535
  |
Correlation:0.030015
  |
Correlation:0.030015
  |
Correlation:0.030015
  |
Correlation:-0.003947
  |
Correlation:-0.078284
  |
Component:385; Jaccard:0.10237
 |
Component:263; Jaccard:0.11502
 |
Component:263; Jaccard:0.12331
 |
Component:361; Jaccard:0.13053
 |
Component:361; Jaccard:0.14162
 |
Component:242; Jaccard:0.14776
 |
Component:242; Jaccard:0.13233
 |
Correlation:-0.10088
  |
Correlation:-0.20267
  |
Correlation:-0.20267
  |
Correlation:-0.003947
  |
Correlation:-0.003947
  |
Correlation:0.030015
  |
Correlation:0.030015
  |
Component:320; Jaccard:0.099952
 |
Component:385; Jaccard:0.11232
 |
Component:385; Jaccard:0.12082
 |
Component:263; Jaccard:0.12804
 |
Component:385; Jaccard:0.12587
 |
Component:385; Jaccard:0.11865
 |
Component:182; Jaccard:0.10601
 |
Correlation:0.14535
  |
Correlation:-0.10088
  |
Correlation:-0.10088
  |
Correlation:-0.20267
  |
Correlation:-0.10088
  |
Correlation:-0.10088
  |
Correlation:0.074667
  |
Component:188; Jaccard:0.097757
 |
Component:188; Jaccard:0.10377
 |
Component:361; Jaccard:0.11717
 |
Component:385; Jaccard:0.12597
 |
Component:263; Jaccard:0.12586
 |
Component:263; Jaccard:0.11706
 |
Component:385; Jaccard:0.10475
 |
Correlation:-0.042124
  |
Correlation:-0.042124
  |
Correlation:-0.003947
  |
Correlation:-0.10088
  |
Correlation:-0.20267
  |
Correlation:-0.20267
  |
Correlation:-0.10088
  |
Component:026; Jaccard:0.094115
 |
Component:361; Jaccard:0.1032
 |
Component:188; Jaccard:0.1089
 |
Component:182; Jaccard:0.11616
 |
Component:182; Jaccard:0.11799
 |
Component:182; Jaccard:0.11463
 |
Component:263; Jaccard:0.10411
 |
Correlation:-0.030144
  |
Correlation:-0.003947
  |
Correlation:-0.042124
  |
Correlation:0.074667
  |
Correlation:0.074667
  |
Correlation:0.074667
  |
Correlation:-0.20267
  |
Cope: 06:neg_REL BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
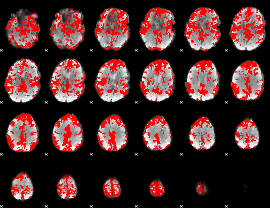 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
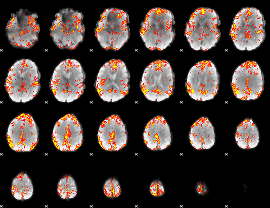 |
GLM Z-score result thresholded by 4
 |
Component:263; Jaccard:0.098643
 |
Component:328; Jaccard:0.11041
 |
Component:328; Jaccard:0.12841
 |
Component:328; Jaccard:0.14775
 |
Component:328; Jaccard:0.17113
 |
Component:328; Jaccard:0.19248
 |
Component:328; Jaccard:0.21202
 |
Correlation:0.64488
  |
Correlation:0.31433
  |
Correlation:0.31433
  |
Correlation:0.31433
  |
Correlation:0.31433
  |
Correlation:0.31433
  |
Correlation:0.31433
  |
Component:328; Jaccard:0.095225
 |
Component:263; Jaccard:0.10927
 |
Component:168; Jaccard:0.12337
 |
Component:168; Jaccard:0.13941
 |
Component:168; Jaccard:0.15553
 |
Component:350; Jaccard:0.173
 |
Component:350; Jaccard:0.19123
 |
Correlation:0.31433
  |
Correlation:0.64488
  |
Correlation:0.23174
  |
Correlation:0.23174
  |
Correlation:0.23174
  |
Correlation:0.28116
  |
Correlation:0.28116
  |
Component:168; Jaccard:0.094553
 |
Component:168; Jaccard:0.10763
 |
Component:385; Jaccard:0.12073
 |
Component:385; Jaccard:0.13405
 |
Component:350; Jaccard:0.15238
 |
Component:168; Jaccard:0.1704
 |
Component:168; Jaccard:0.1798
 |
Correlation:0.23174
  |
Correlation:0.23174
  |
Correlation:0.43809
  |
Correlation:0.43809
  |
Correlation:0.28116
  |
Correlation:0.23174
  |
Correlation:0.23174
  |
Component:385; Jaccard:0.093814
 |
Component:385; Jaccard:0.10673
 |
Component:263; Jaccard:0.1198
 |
Component:350; Jaccard:0.13362
 |
Component:385; Jaccard:0.14592
 |
Component:385; Jaccard:0.15482
 |
Component:385; Jaccard:0.15811
 |
Correlation:0.43809
  |
Correlation:0.43809
  |
Correlation:0.64488
  |
Correlation:0.28116
  |
Correlation:0.43809
  |
Correlation:0.43809
  |
Correlation:0.43809
  |
Component:253; Jaccard:0.091409
 |
Component:026; Jaccard:0.10391
 |
Component:026; Jaccard:0.1174
 |
Component:263; Jaccard:0.13052
 |
Component:026; Jaccard:0.14221
 |
Component:026; Jaccard:0.15112
 |
Component:026; Jaccard:0.15614
 |
Correlation:0.4798
  |
Correlation:0.3913
  |
Correlation:0.3913
  |
Correlation:0.64488
  |
Correlation:0.3913
  |
Correlation:0.3913
  |
Correlation:0.3913
  |
Component:026; Jaccard:0.091195
 |
Component:253; Jaccard:0.10239
 |
Component:350; Jaccard:0.11553
 |
Component:026; Jaccard:0.13047
 |
Component:263; Jaccard:0.13981
 |
Component:263; Jaccard:0.14682
 |
Component:361; Jaccard:0.15585
 |
Correlation:0.3913
  |
Correlation:0.4798
  |
Correlation:0.28116
  |
Correlation:0.3913
  |
Correlation:0.64488
  |
Correlation:0.64488
  |
Correlation:0.46344
  |
Component:138; Jaccard:0.090465
 |
Component:138; Jaccard:0.10121
 |
Component:253; Jaccard:0.11388
 |
Component:253; Jaccard:0.12417
 |
Component:253; Jaccard:0.13252
 |
Component:361; Jaccard:0.14243
 |
Component:263; Jaccard:0.14929
 |
Correlation:0.26675
  |
Correlation:0.26675
  |
Correlation:0.4798
  |
Correlation:0.4798
  |
Correlation:0.4798
  |
Correlation:0.46344
  |
Correlation:0.64488
  |
Component:156; Jaccard:0.086827
 |
Component:350; Jaccard:0.099348
 |
Component:138; Jaccard:0.11193
 |
Component:138; Jaccard:0.12263
 |
Component:138; Jaccard:0.13168
 |
Component:156; Jaccard:0.13981
 |
Component:156; Jaccard:0.14903
 |
Correlation:0.28701
  |
Correlation:0.28116
  |
Correlation:0.26675
  |
Correlation:0.26675
  |
Correlation:0.26675
  |
Correlation:0.28701
  |
Correlation:0.28701
  |
Component:216; Jaccard:0.086662
 |
Component:156; Jaccard:0.097024
 |
Component:156; Jaccard:0.10835
 |
Component:242; Jaccard:0.11925
 |
Component:242; Jaccard:0.13034
 |
Component:138; Jaccard:0.13859
 |
Component:320; Jaccard:0.14771
 |
Correlation:0.431
  |
Correlation:0.28701
  |
Correlation:0.28701
  |
Correlation:0.24943
  |
Correlation:0.24943
  |
Correlation:0.26675
  |
Correlation:0.15788
  |
Component:350; Jaccard:0.086297
 |
Component:242; Jaccard:0.095534
 |
Component:242; Jaccard:0.10731
 |
Component:156; Jaccard:0.11885
 |
Component:156; Jaccard:0.13004
 |
Component:242; Jaccard:0.13858
 |
Component:242; Jaccard:0.14162
 |
Correlation:0.28116
  |
Correlation:0.24943
  |
Correlation:0.24943
  |
Correlation:0.28701
  |
Correlation:0.28701
  |
Correlation:0.24943
  |
Correlation:0.24943
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































