Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
Cope: 01:FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
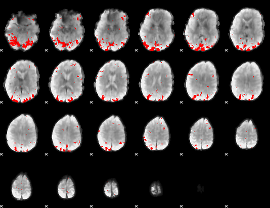 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
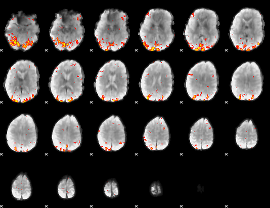 |
GLM Z-score result thresholded by 3.5
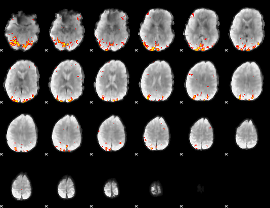 |
GLM Z-score result thresholded by 4
 |
Component:371; Jaccard:0.13548
 |
Component:371; Jaccard:0.17331
 |
Component:209; Jaccard:0.21236
 |
Component:209; Jaccard:0.23551
 |
Component:209; Jaccard:0.23484
 |
Component:209; Jaccard:0.21709
 |
Component:209; Jaccard:0.18846
 |
Correlation:0.66399
 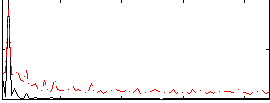 |
Correlation:0.66399
  |
Correlation:0.20609
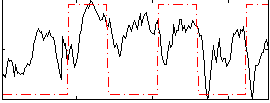  |
Correlation:0.20609
  |
Correlation:0.20609
  |
Correlation:0.20609
  |
Correlation:0.20609
  |
Component:209; Jaccard:0.12723
 |
Component:209; Jaccard:0.17177
 |
Component:371; Jaccard:0.20571
 |
Component:371; Jaccard:0.22064
 |
Component:371; Jaccard:0.20843
 |
Component:228; Jaccard:0.20209
 |
Component:228; Jaccard:0.18573
 |
Correlation:0.20609
  |
Correlation:0.20609
  |
Correlation:0.66399
  |
Correlation:0.66399
  |
Correlation:0.66399
  |
Correlation:0.21455
  |
Correlation:0.21455
  |
Component:316; Jaccard:0.10631
 |
Component:316; Jaccard:0.13686
 |
Component:316; Jaccard:0.16494
 |
Component:228; Jaccard:0.18907
 |
Component:228; Jaccard:0.20213
 |
Component:371; Jaccard:0.18321
 |
Component:371; Jaccard:0.15965
 |
Correlation:-0.013776
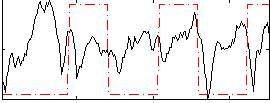  |
Correlation:-0.013776
  |
Correlation:-0.013776
  |
Correlation:0.21455
  |
Correlation:0.21455
  |
Correlation:0.66399
  |
Correlation:0.66399
  |
Component:370; Jaccard:0.099252
 |
Component:370; Jaccard:0.12922
 |
Component:228; Jaccard:0.16183
 |
Component:316; Jaccard:0.18635
 |
Component:316; Jaccard:0.18738
 |
Component:370; Jaccard:0.17719
 |
Component:370; Jaccard:0.15737
 |
Correlation:0.56901
  |
Correlation:0.56901
  |
Correlation:0.21455
  |
Correlation:-0.013776
  |
Correlation:-0.013776
  |
Correlation:0.56901
  |
Correlation:0.56901
  |
Component:228; Jaccard:0.091679
 |
Component:228; Jaccard:0.12614
 |
Component:370; Jaccard:0.16009
 |
Component:370; Jaccard:0.18264
 |
Component:370; Jaccard:0.18601
 |
Component:316; Jaccard:0.17363
 |
Component:316; Jaccard:0.15388
 |
Correlation:0.21455
  |
Correlation:0.21455
  |
Correlation:0.56901
  |
Correlation:0.56901
  |
Correlation:0.56901
  |
Correlation:-0.013776
  |
Correlation:-0.013776
  |
Component:326; Jaccard:0.087721
 |
Component:326; Jaccard:0.10192
 |
Component:326; Jaccard:0.10973
 |
Component:326; Jaccard:0.1106
 |
Component:326; Jaccard:0.10151
 |
Component:326; Jaccard:0.090747
 |
Component:326; Jaccard:0.079293
 |
Correlation:0.23408
 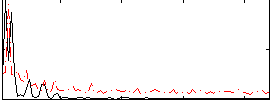 |
Correlation:0.23408
  |
Correlation:0.23408
  |
Correlation:0.23408
  |
Correlation:0.23408
  |
Correlation:0.23408
  |
Correlation:0.23408
  |
Component:072; Jaccard:0.077115
 |
Component:072; Jaccard:0.090402
 |
Component:072; Jaccard:0.10008
 |
Component:072; Jaccard:0.10166
 |
Component:072; Jaccard:0.093968
 |
Component:150; Jaccard:0.084574
 |
Component:150; Jaccard:0.072096
 |
Correlation:0.057125
 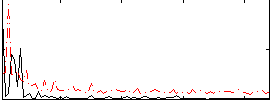 |
Correlation:0.057125
  |
Correlation:0.057125
  |
Correlation:0.057125
  |
Correlation:0.057125
  |
Correlation:0.39325
  |
Correlation:0.39325
  |
Component:150; Jaccard:0.073069
 |
Component:150; Jaccard:0.082816
 |
Component:150; Jaccard:0.088334
 |
Component:150; Jaccard:0.09193
 |
Component:150; Jaccard:0.089667
 |
Component:072; Jaccard:0.081103
 |
Component:072; Jaccard:0.066775
 |
Correlation:0.39325
  |
Correlation:0.39325
  |
Correlation:0.39325
  |
Correlation:0.39325
  |
Correlation:0.39325
  |
Correlation:0.057125
  |
Correlation:0.057125
  |
Component:081; Jaccard:0.071695
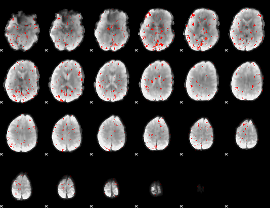 |
Component:081; Jaccard:0.075366
 |
Component:080; Jaccard:0.079063
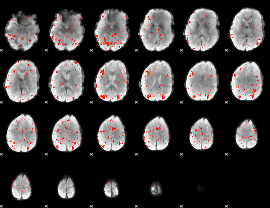 |
Component:080; Jaccard:0.077384
 |
Component:344; Jaccard:0.072052
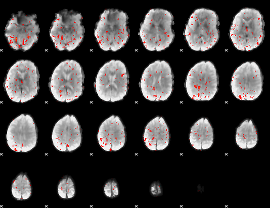 |
Component:344; Jaccard:0.064597
 |
Component:344; Jaccard:0.057101
 |
Correlation:0.11046
  |
Correlation:0.11046
  |
Correlation:0.35694
 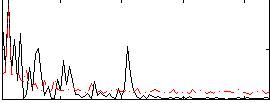 |
Correlation:0.35694
  |
Correlation:0.1448
 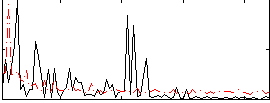 |
Correlation:0.1448
  |
Correlation:0.1448
  |
Component:115; Jaccard:0.071137
 |
Component:080; Jaccard:0.074973
 |
Component:128; Jaccard:0.074869
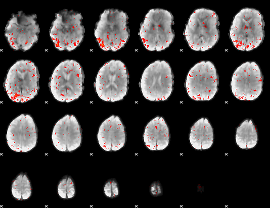 |
Component:344; Jaccard:0.074458
 |
Component:080; Jaccard:0.069657
 |
Component:080; Jaccard:0.058459
 |
Component:230; Jaccard:0.048184
 |
Correlation:0.27677
  |
Correlation:0.35694
  |
Correlation:0.14389
  |
Correlation:0.1448
  |
Correlation:0.35694
  |
Correlation:0.35694
  |
Correlation:0.32741
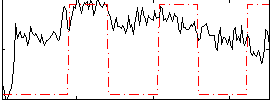  |
Cope: 02:SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
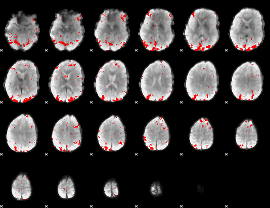 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
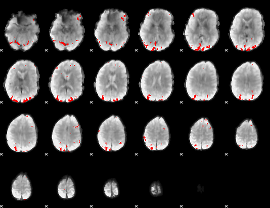 |
GLM binary result thresholded by 4
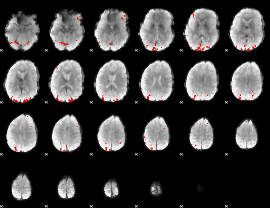 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
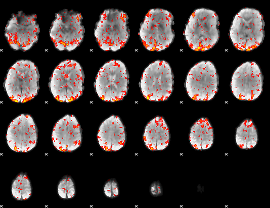 |
GLM Z-score result thresholded by 2
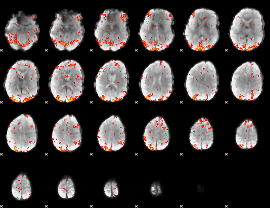 |
GLM Z-score result thresholded by 2.5
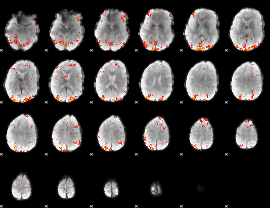 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
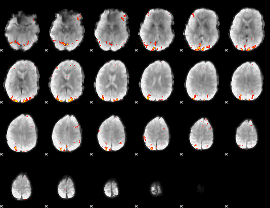 |
GLM Z-score result thresholded by 4
 |
Component:209; Jaccard:0.14121
 |
Component:209; Jaccard:0.19092
 |
Component:209; Jaccard:0.23358
 |
Component:209; Jaccard:0.25232
 |
Component:209; Jaccard:0.23376
 |
Component:209; Jaccard:0.19902
 |
Component:228; Jaccard:0.17294
 |
Correlation:-0.060801
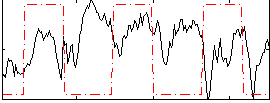  |
Correlation:-0.060801
  |
Correlation:-0.060801
  |
Correlation:-0.060801
  |
Correlation:-0.060801
  |
Correlation:-0.060801
  |
Correlation:-0.053844
  |
Component:316; Jaccard:0.11869
 |
Component:316; Jaccard:0.15966
 |
Component:316; Jaccard:0.19574
 |
Component:316; Jaccard:0.21071
 |
Component:228; Jaccard:0.21716
 |
Component:228; Jaccard:0.1971
 |
Component:209; Jaccard:0.15371
 |
Correlation:0.17762
  |
Correlation:0.17762
  |
Correlation:0.17762
  |
Correlation:0.17762
  |
Correlation:-0.053844
  |
Correlation:-0.053844
  |
Correlation:-0.060801
  |
Component:228; Jaccard:0.10046
 |
Component:228; Jaccard:0.13889
 |
Component:228; Jaccard:0.17922
 |
Component:228; Jaccard:0.20865
 |
Component:316; Jaccard:0.20388
 |
Component:316; Jaccard:0.18149
 |
Component:316; Jaccard:0.15071
 |
Correlation:-0.053844
  |
Correlation:-0.053844
  |
Correlation:-0.053844
  |
Correlation:-0.053844
  |
Correlation:0.17762
  |
Correlation:0.17762
  |
Correlation:0.17762
  |
Component:376; Jaccard:0.09274
 |
Component:072; Jaccard:0.10112
 |
Component:072; Jaccard:0.10889
 |
Component:072; Jaccard:0.10676
 |
Component:072; Jaccard:0.093786
 |
Component:072; Jaccard:0.074383
 |
Component:072; Jaccard:0.05953
 |
Correlation:0.18568
  |
Correlation:0.17024
  |
Correlation:0.17024
  |
Correlation:0.17024
  |
Correlation:0.17024
  |
Correlation:0.17024
  |
Correlation:0.17024
  |
Component:072; Jaccard:0.085254
 |
Component:376; Jaccard:0.09693
 |
Component:376; Jaccard:0.093027
 |
Component:370; Jaccard:0.079634
 |
Component:370; Jaccard:0.072931
 |
Component:370; Jaccard:0.059842
 |
Component:370; Jaccard:0.04657
 |
Correlation:0.17024
  |
Correlation:0.18568
  |
Correlation:0.18568
  |
Correlation:-0.29228
  |
Correlation:-0.29228
  |
Correlation:-0.29228
  |
Correlation:-0.29228
  |
Component:115; Jaccard:0.077731
 |
Component:191; Jaccard:0.079925
 |
Component:191; Jaccard:0.078003
 |
Component:376; Jaccard:0.077067
 |
Component:191; Jaccard:0.059348
 |
Component:191; Jaccard:0.048516
 |
Component:191; Jaccard:0.037276
 |
Correlation:-0.16807
  |
Correlation:0.066101
  |
Correlation:0.066101
  |
Correlation:0.18568
  |
Correlation:0.066101
  |
Correlation:0.066101
  |
Correlation:0.066101
  |
Component:191; Jaccard:0.077312
 |
Component:115; Jaccard:0.079156
 |
Component:370; Jaccard:0.077949
 |
Component:191; Jaccard:0.068487
 |
Component:376; Jaccard:0.055416
 |
Component:230; Jaccard:0.042906
 |
Component:230; Jaccard:0.034045
 |
Correlation:0.066101
  |
Correlation:-0.16807
  |
Correlation:-0.29228
  |
Correlation:0.066101
  |
Correlation:0.18568
  |
Correlation:-0.055979
  |
Correlation:-0.055979
  |
Component:299; Jaccard:0.066054
 |
Component:370; Jaccard:0.071152
 |
Component:115; Jaccard:0.072145
 |
Component:230; Jaccard:0.060039
 |
Component:230; Jaccard:0.053935
 |
Component:344; Jaccard:0.038832
 |
Component:344; Jaccard:0.030815
 |
Correlation:-0.035563
  |
Correlation:-0.29228
  |
Correlation:-0.16807
  |
Correlation:-0.055979
  |
Correlation:-0.055979
  |
Correlation:-0.19245
  |
Correlation:-0.19245
  |
Component:283; Jaccard:0.065316
 |
Component:283; Jaccard:0.070817
 |
Component:283; Jaccard:0.068692
 |
Component:115; Jaccard:0.059504
 |
Component:344; Jaccard:0.047456
 |
Component:376; Jaccard:0.037429
 |
Component:390; Jaccard:0.030179
 |
Correlation:-0.0073514
  |
Correlation:-0.0073514
  |
Correlation:-0.0073514
  |
Correlation:-0.16807
  |
Correlation:-0.19245
  |
Correlation:0.18568
  |
Correlation:0.23013
 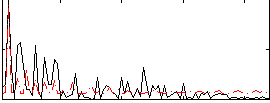 |
Component:370; Jaccard:0.061203
 |
Component:299; Jaccard:0.068412
 |
Component:299; Jaccard:0.06636
 |
Component:283; Jaccard:0.05902
 |
Component:283; Jaccard:0.046144
 |
Component:080; Jaccard:0.036621
 |
Component:080; Jaccard:0.026409
 |
Correlation:-0.29228
  |
Correlation:-0.035563
  |
Correlation:-0.035563
  |
Correlation:-0.0073514
  |
Correlation:-0.0073514
  |
Correlation:-0.30534
  |
Correlation:-0.30534
  |
Cope: 03:FACES-SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
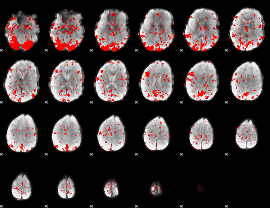 |
GLM binary result thresholded by 2.5
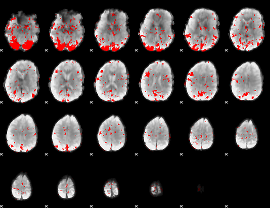 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
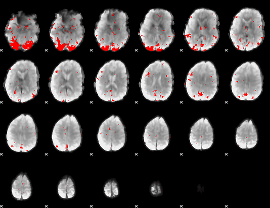 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
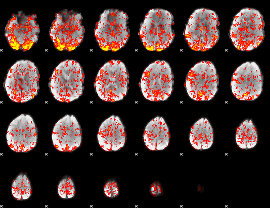 |
GLM Z-score result thresholded by 1.5
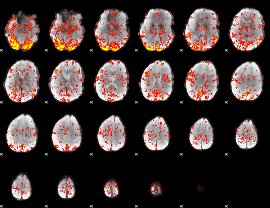 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
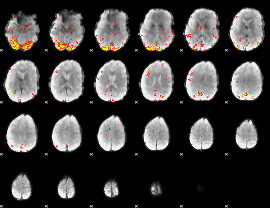 |
GLM Z-score result thresholded by 4
 |
Component:371; Jaccard:0.1322
 |
Component:371; Jaccard:0.17879
 |
Component:371; Jaccard:0.23205
 |
Component:371; Jaccard:0.28132
 |
Component:371; Jaccard:0.30953
 |
Component:371; Jaccard:0.30956
 |
Component:371; Jaccard:0.2884
 |
Correlation:0.7021
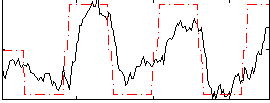 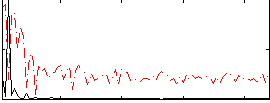 |
Correlation:0.7021
  |
Correlation:0.7021
  |
Correlation:0.7021
  |
Correlation:0.7021
  |
Correlation:0.7021
  |
Correlation:0.7021
  |
Component:056; Jaccard:0.122
 |
Component:056; Jaccard:0.16077
 |
Component:056; Jaccard:0.2008
 |
Component:056; Jaccard:0.2286
 |
Component:056; Jaccard:0.24241
 |
Component:056; Jaccard:0.23984
 |
Component:056; Jaccard:0.23011
 |
Correlation:0.64535
  |
Correlation:0.64535
  |
Correlation:0.64535
  |
Correlation:0.64535
  |
Correlation:0.64535
  |
Correlation:0.64535
  |
Correlation:0.64535
  |
Component:326; Jaccard:0.10339
 |
Component:326; Jaccard:0.12841
 |
Component:326; Jaccard:0.15386
 |
Component:370; Jaccard:0.16802
 |
Component:370; Jaccard:0.1821
 |
Component:370; Jaccard:0.18371
 |
Component:370; Jaccard:0.17214
 |
Correlation:0.17703
 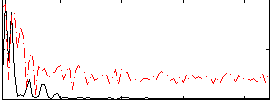 |
Correlation:0.17703
  |
Correlation:0.17703
  |
Correlation:0.46717
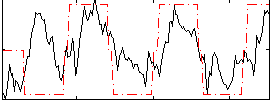 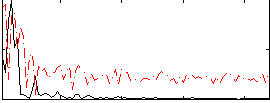 |
Correlation:0.46717
  |
Correlation:0.46717
  |
Correlation:0.46717
  |
Component:150; Jaccard:0.09201
 |
Component:150; Jaccard:0.1141
 |
Component:370; Jaccard:0.14024
 |
Component:326; Jaccard:0.16567
 |
Component:326; Jaccard:0.15981
 |
Component:326; Jaccard:0.14499
 |
Component:326; Jaccard:0.12768
 |
Correlation:0.41635
 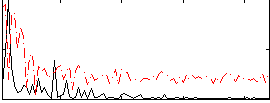 |
Correlation:0.41635
  |
Correlation:0.46717
  |
Correlation:0.17703
  |
Correlation:0.17703
  |
Correlation:0.17703
  |
Correlation:0.17703
  |
Component:370; Jaccard:0.085576
 |
Component:370; Jaccard:0.11061
 |
Component:150; Jaccard:0.13605
 |
Component:150; Jaccard:0.14649
 |
Component:150; Jaccard:0.14741
 |
Component:150; Jaccard:0.13717
 |
Component:150; Jaccard:0.12503
 |
Correlation:0.46717
  |
Correlation:0.46717
  |
Correlation:0.41635
  |
Correlation:0.41635
  |
Correlation:0.41635
  |
Correlation:0.41635
  |
Correlation:0.41635
  |
Component:110; Jaccard:0.078275
 |
Component:110; Jaccard:0.088382
 |
Component:110; Jaccard:0.092851
 |
Component:322; Jaccard:0.092261
 |
Component:322; Jaccard:0.085302
 |
Component:208; Jaccard:0.078955
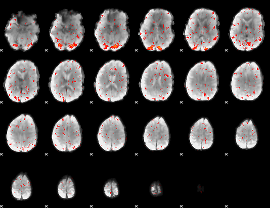 |
Component:208; Jaccard:0.072966
 |
Correlation:0.27079
  |
Correlation:0.27079
  |
Correlation:0.27079
  |
Correlation:0.19975
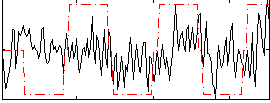  |
Correlation:0.19975
  |
Correlation:0.35638
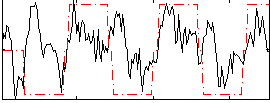  |
Correlation:0.35638
  |
Component:322; Jaccard:0.076503
 |
Component:322; Jaccard:0.086518
 |
Component:322; Jaccard:0.091583
 |
Component:110; Jaccard:0.08858
 |
Component:208; Jaccard:0.083786
 |
Component:322; Jaccard:0.073396
 |
Component:322; Jaccard:0.061952
 |
Correlation:0.19975
  |
Correlation:0.19975
  |
Correlation:0.19975
  |
Correlation:0.27079
  |
Correlation:0.35638
  |
Correlation:0.19975
  |
Correlation:0.19975
  |
Component:187; Jaccard:0.074669
 |
Component:187; Jaccard:0.081006
 |
Component:208; Jaccard:0.085513
 |
Component:208; Jaccard:0.084271
 |
Component:110; Jaccard:0.07686
 |
Component:110; Jaccard:0.064452
 |
Component:110; Jaccard:0.052976
 |
Correlation:0.12176
  |
Correlation:0.12176
  |
Correlation:0.35638
  |
Correlation:0.35638
  |
Correlation:0.27079
  |
Correlation:0.27079
  |
Correlation:0.27079
  |
Component:208; Jaccard:0.069843
 |
Component:208; Jaccard:0.079478
 |
Component:187; Jaccard:0.085228
 |
Component:187; Jaccard:0.082757
 |
Component:187; Jaccard:0.073702
 |
Component:187; Jaccard:0.06131
 |
Component:210; Jaccard:0.05291
 |
Correlation:0.35638
  |
Correlation:0.35638
  |
Correlation:0.12176
  |
Correlation:0.12176
  |
Correlation:0.12176
  |
Correlation:0.12176
  |
Correlation:0.30445
 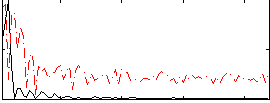 |
Component:145; Jaccard:0.062402
 |
Component:145; Jaccard:0.066811
 |
Component:260; Jaccard:0.070675
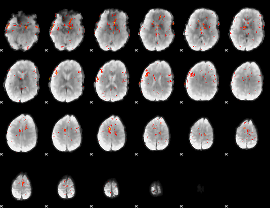 |
Component:260; Jaccard:0.076047
 |
Component:260; Jaccard:0.068471
 |
Component:209; Jaccard:0.056343
 |
Component:187; Jaccard:0.051318
 |
Correlation:0.23569
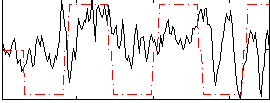  |
Correlation:0.23569
  |
Correlation:0.38169
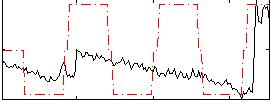  |
Correlation:0.38169
  |
Correlation:0.38169
  |
Correlation:0.14477
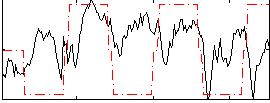 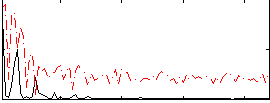 |
Correlation:0.12176
  |
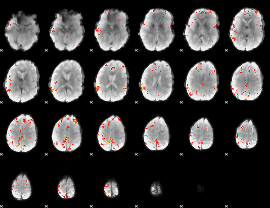
Cope: 04:neg_FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
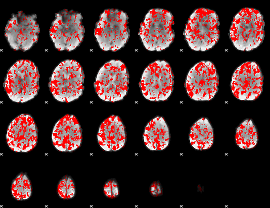 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
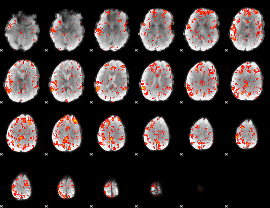 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:345; Jaccard:0.11169
 |
Component:345; Jaccard:0.14205
 |
Component:345; Jaccard:0.16858
 |
Component:345; Jaccard:0.17979
 |
Component:345; Jaccard:0.17108
 |
Component:345; Jaccard:0.14466
 |
Component:345; Jaccard:0.10946
 |
Correlation:-0.016139
  |
Correlation:-0.016139
  |
Correlation:-0.016139
  |
Correlation:-0.016139
  |
Correlation:-0.016139
  |
Correlation:-0.016139
  |
Correlation:-0.016139
  |
Component:246; Jaccard:0.10401
 |
Component:246; Jaccard:0.13268
 |
Component:142; Jaccard:0.15937
 |
Component:142; Jaccard:0.17296
 |
Component:142; Jaccard:0.16331
 |
Component:142; Jaccard:0.14213
 |
Component:142; Jaccard:0.10504
 |
Correlation:-0.092446
  |
Correlation:-0.092446
  |
Correlation:0.24442
  |
Correlation:0.24442
  |
Correlation:0.24442
  |
Correlation:0.24442
  |
Correlation:0.24442
  |
Component:142; Jaccard:0.10155
 |
Component:142; Jaccard:0.13037
 |
Component:246; Jaccard:0.15876
 |
Component:246; Jaccard:0.17088
 |
Component:246; Jaccard:0.16313
 |
Component:246; Jaccard:0.13831
 |
Component:246; Jaccard:0.10409
 |
Correlation:0.24442
  |
Correlation:0.24442
  |
Correlation:-0.092446
  |
Correlation:-0.092446
  |
Correlation:-0.092446
  |
Correlation:-0.092446
  |
Correlation:-0.092446
  |
Component:309; Jaccard:0.085816
 |
Component:309; Jaccard:0.10064
 |
Component:309; Jaccard:0.11237
 |
Component:309; Jaccard:0.11169
 |
Component:312; Jaccard:0.10867
 |
Component:312; Jaccard:0.091711
 |
Component:312; Jaccard:0.067897
 |
Correlation:-0.037077
  |
Correlation:-0.037077
  |
Correlation:-0.037077
  |
Correlation:-0.037077
  |
Correlation:-0.20036
  |
Correlation:-0.20036
  |
Correlation:-0.20036
  |
Component:254; Jaccard:0.082328
 |
Component:254; Jaccard:0.096237
 |
Component:254; Jaccard:0.10623
 |
Component:312; Jaccard:0.11055
 |
Component:236; Jaccard:0.10096
 |
Component:236; Jaccard:0.082472
 |
Component:236; Jaccard:0.058916
 |
Correlation:0.099449
  |
Correlation:0.099449
  |
Correlation:0.099449
  |
Correlation:-0.20036
  |
Correlation:0.17926
  |
Correlation:0.17926
  |
Correlation:0.17926
  |
Component:312; Jaccard:0.08026
 |
Component:312; Jaccard:0.094051
 |
Component:312; Jaccard:0.10539
 |
Component:236; Jaccard:0.10686
 |
Component:309; Jaccard:0.09995
 |
Component:309; Jaccard:0.081673
 |
Component:309; Jaccard:0.058232
 |
Correlation:-0.20036
  |
Correlation:-0.20036
  |
Correlation:-0.20036
  |
Correlation:0.17926
  |
Correlation:-0.037077
  |
Correlation:-0.037077
  |
Correlation:-0.037077
  |
Component:156; Jaccard:0.076157
 |
Component:236; Jaccard:0.086134
 |
Component:236; Jaccard:0.098699
 |
Component:254; Jaccard:0.10491
 |
Component:254; Jaccard:0.0893
 |
Component:254; Jaccard:0.06833
 |
Component:254; Jaccard:0.046846
 |
Correlation:-0.19687
  |
Correlation:0.17926
  |
Correlation:0.17926
  |
Correlation:0.099449
  |
Correlation:0.099449
  |
Correlation:0.099449
  |
Correlation:0.099449
  |
Component:236; Jaccard:0.073271
 |
Component:156; Jaccard:0.083439
 |
Component:156; Jaccard:0.088085
 |
Component:282; Jaccard:0.086299
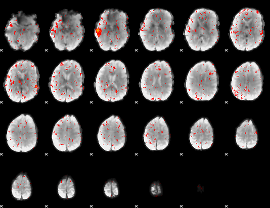 |
Component:282; Jaccard:0.078021
 |
Component:282; Jaccard:0.060068
 |
Component:206; Jaccard:0.044536
 |
Correlation:0.17926
  |
Correlation:-0.19687
  |
Correlation:-0.19687
  |
Correlation:0.30051
  |
Correlation:0.30051
  |
Correlation:0.30051
  |
Correlation:0.053824
  |
Component:244; Jaccard:0.070605
 |
Component:282; Jaccard:0.079129
 |
Component:282; Jaccard:0.084657
 |
Component:156; Jaccard:0.084362
 |
Component:206; Jaccard:0.074277
 |
Component:206; Jaccard:0.059281
 |
Component:282; Jaccard:0.044532
 |
Correlation:0.081608
  |
Correlation:0.30051
  |
Correlation:0.30051
  |
Correlation:-0.19687
  |
Correlation:0.053824
  |
Correlation:0.053824
  |
Correlation:0.30051
  |
Component:282; Jaccard:0.070363
 |
Component:050; Jaccard:0.078751
 |
Component:050; Jaccard:0.083195
 |
Component:206; Jaccard:0.083543
 |
Component:156; Jaccard:0.073393
 |
Component:244; Jaccard:0.058086
 |
Component:050; Jaccard:0.040088
 |
Correlation:0.30051
  |
Correlation:0.06638
  |
Correlation:0.06638
  |
Correlation:0.053824
  |
Correlation:-0.19687
  |
Correlation:0.081608
  |
Correlation:0.06638
  |
Cope: 05:neg_SHAPES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
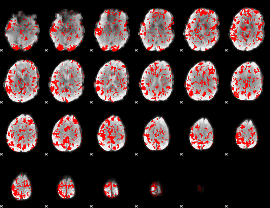 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
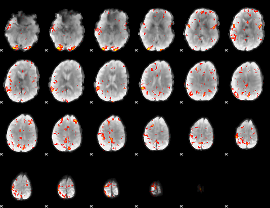 |
GLM Z-score result thresholded by 3.5
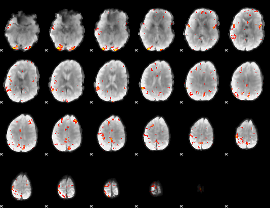 |
GLM Z-score result thresholded by 4
 |
Component:246; Jaccard:0.095978
 |
Component:246; Jaccard:0.12056
 |
Component:246; Jaccard:0.14026
 |
Component:246; Jaccard:0.14593
 |
Component:246; Jaccard:0.13695
 |
Component:056; Jaccard:0.11688
 |
Component:056; Jaccard:0.099763
 |
Correlation:0.21709
  |
Correlation:0.21709
  |
Correlation:0.21709
  |
Correlation:0.21709
  |
Correlation:0.21709
  |
Correlation:0.62942
  |
Correlation:0.62942
  |
Component:345; Jaccard:0.089644
 |
Component:345; Jaccard:0.10851
 |
Component:345; Jaccard:0.12282
 |
Component:056; Jaccard:0.13066
 |
Component:056; Jaccard:0.12866
 |
Component:246; Jaccard:0.10952
 |
Component:246; Jaccard:0.081484
 |
Correlation:0.16443
  |
Correlation:0.16443
  |
Correlation:0.16443
  |
Correlation:0.62942
  |
Correlation:0.62942
  |
Correlation:0.21709
  |
Correlation:0.21709
  |
Component:056; Jaccard:0.088596
 |
Component:056; Jaccard:0.10748
 |
Component:056; Jaccard:0.1223
 |
Component:345; Jaccard:0.12902
 |
Component:345; Jaccard:0.11902
 |
Component:236; Jaccard:0.099964
 |
Component:236; Jaccard:0.075881
 |
Correlation:0.62942
  |
Correlation:0.62942
  |
Correlation:0.62942
  |
Correlation:0.16443
  |
Correlation:0.16443
  |
Correlation:-0.052072
  |
Correlation:-0.052072
  |
Component:244; Jaccard:0.082277
 |
Component:244; Jaccard:0.095045
 |
Component:312; Jaccard:0.10935
 |
Component:236; Jaccard:0.11747
 |
Component:236; Jaccard:0.11432
 |
Component:345; Jaccard:0.095312
 |
Component:345; Jaccard:0.070514
 |
Correlation:-0.0038583
  |
Correlation:-0.0038583
  |
Correlation:0.22674
  |
Correlation:-0.052072
  |
Correlation:-0.052072
  |
Correlation:0.16443
  |
Correlation:0.16443
  |
Component:312; Jaccard:0.079619
 |
Component:312; Jaccard:0.09479
 |
Component:236; Jaccard:0.10748
 |
Component:312; Jaccard:0.11639
 |
Component:312; Jaccard:0.10942
 |
Component:312; Jaccard:0.090485
 |
Component:312; Jaccard:0.06751
 |
Correlation:0.22674
  |
Correlation:0.22674
  |
Correlation:-0.052072
  |
Correlation:0.22674
  |
Correlation:0.22674
  |
Correlation:0.22674
  |
Correlation:0.22674
  |
Component:142; Jaccard:0.077442
 |
Component:236; Jaccard:0.092193
 |
Component:244; Jaccard:0.1063
 |
Component:254; Jaccard:0.11213
 |
Component:254; Jaccard:0.10624
 |
Component:254; Jaccard:0.087111
 |
Component:254; Jaccard:0.066985
 |
Correlation:-0.26718
  |
Correlation:-0.052072
  |
Correlation:-0.0038583
  |
Correlation:0.064886
  |
Correlation:0.064886
  |
Correlation:0.064886
  |
Correlation:0.064886
  |
Component:050; Jaccard:0.076797
 |
Component:142; Jaccard:0.091449
 |
Component:254; Jaccard:0.10502
 |
Component:244; Jaccard:0.10674
 |
Component:244; Jaccard:0.099376
 |
Component:244; Jaccard:0.082728
 |
Component:244; Jaccard:0.063525
 |
Correlation:0.12396
  |
Correlation:-0.26718
  |
Correlation:0.064886
  |
Correlation:-0.0038583
  |
Correlation:-0.0038583
  |
Correlation:-0.0038583
  |
Correlation:-0.0038583
  |
Component:254; Jaccard:0.076634
 |
Component:254; Jaccard:0.091183
 |
Component:050; Jaccard:0.10183
 |
Component:142; Jaccard:0.10592
 |
Component:050; Jaccard:0.095032
 |
Component:050; Jaccard:0.07693
 |
Component:050; Jaccard:0.057917
 |
Correlation:0.064886
  |
Correlation:0.064886
  |
Correlation:0.12396
  |
Correlation:-0.26718
  |
Correlation:0.12396
  |
Correlation:0.12396
  |
Correlation:0.12396
  |
Component:236; Jaccard:0.075877
 |
Component:050; Jaccard:0.090927
 |
Component:142; Jaccard:0.101
 |
Component:050; Jaccard:0.10363
 |
Component:142; Jaccard:0.094629
 |
Component:142; Jaccard:0.075856
 |
Component:142; Jaccard:0.054079
 |
Correlation:-0.052072
  |
Correlation:0.12396
  |
Correlation:-0.26718
  |
Correlation:0.12396
  |
Correlation:-0.26718
  |
Correlation:-0.26718
  |
Correlation:-0.26718
  |
Component:309; Jaccard:0.074292
 |
Component:309; Jaccard:0.085777
 |
Component:309; Jaccard:0.093149
 |
Component:309; Jaccard:0.094426
 |
Component:309; Jaccard:0.087808
 |
Component:309; Jaccard:0.069055
 |
Component:309; Jaccard:0.051204
 |
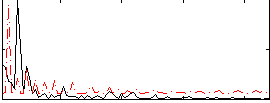
Correlation:0.081184
  |
Correlation:0.081184
  |
Correlation:0.081184
  |
Correlation:0.081184
  |
Correlation:0.081184
  |
Correlation:0.081184
  |
Correlation:0.081184
  |
Cope: 06:SHAPES-FACES BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
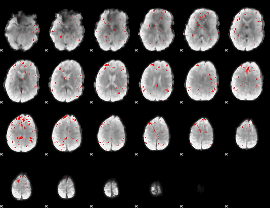
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
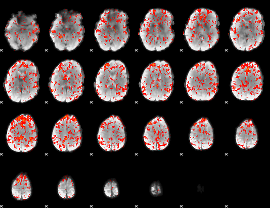 |
GLM Z-score result thresholded by 2
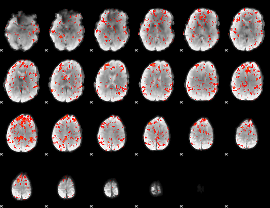 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:142; Jaccard:0.085117
 |
Component:142; Jaccard:0.096299
 |
Component:142; Jaccard:0.10208
 |
Component:142; Jaccard:0.089901
 |
Component:313; Jaccard:0.068364
 |
Component:313; Jaccard:0.044007
 |
Component:142; Jaccard:0.021957
 |
Correlation:0.2775
  |
Correlation:0.2775
  |
Correlation:0.2775
  |
Correlation:0.2775
  |
Correlation:0.07333
  |
Correlation:0.07333
  |
Correlation:0.2775
  |
Component:156; Jaccard:0.079463
 |
Component:156; Jaccard:0.084782
 |
Component:313; Jaccard:0.087201
 |
Component:313; Jaccard:0.083515
 |
Component:007; Jaccard:0.065617
 |
Component:142; Jaccard:0.041613
 |
Component:313; Jaccard:0.021148
 |
Correlation:-0.13461
  |
Correlation:-0.13461
  |
Correlation:0.07333
  |
Correlation:0.07333
  |
Correlation:0.28966
  |
Correlation:0.2775
  |
Correlation:0.07333
  |
Component:345; Jaccard:0.077149
 |
Component:345; Jaccard:0.084401
 |
Component:054; Jaccard:0.086963
 |
Component:007; Jaccard:0.079516
 |
Component:142; Jaccard:0.065435
 |
Component:007; Jaccard:0.037672
 |
Component:007; Jaccard:0.016871
 |
Correlation:-0.097945
  |
Correlation:-0.097945
  |
Correlation:0.41688
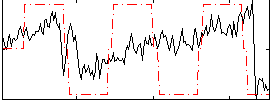  |
Correlation:0.28966
  |
Correlation:0.2775
  |
Correlation:0.28966
  |
Correlation:0.28966
  |
Component:168; Jaccard:0.075862
 |
Component:054; Jaccard:0.081716
 |
Component:345; Jaccard:0.086742
 |
Component:054; Jaccard:0.078542
 |
Component:054; Jaccard:0.058848
 |
Component:168; Jaccard:0.035223
 |
Component:054; Jaccard:0.016351
 |
Correlation:0.014366
  |
Correlation:0.41688
  |
Correlation:-0.097945
  |
Correlation:0.41688
  |
Correlation:0.41688
  |
Correlation:0.014366
  |
Correlation:0.41688
  |
Component:299; Jaccard:0.073462
 |
Component:168; Jaccard:0.080886
 |
Component:007; Jaccard:0.083483
 |
Component:345; Jaccard:0.077582
 |
Component:345; Jaccard:0.05833
 |
Component:054; Jaccard:0.034302
 |
Component:345; Jaccard:0.016228
 |
Correlation:0.0042478
  |
Correlation:0.014366
  |
Correlation:0.28966
  |
Correlation:-0.097945
  |
Correlation:-0.097945
  |
Correlation:0.41688
  |
Correlation:-0.097945
  |
Component:054; Jaccard:0.073018
 |
Component:313; Jaccard:0.080475
 |
Component:156; Jaccard:0.083367
 |
Component:168; Jaccard:0.073793
 |
Component:168; Jaccard:0.057137
 |
Component:345; Jaccard:0.03213
 |
Component:364; Jaccard:0.014744
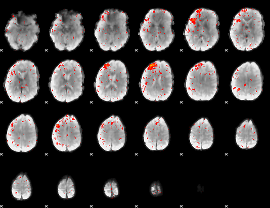 |
Correlation:0.41688
  |
Correlation:0.07333
  |
Correlation:-0.13461
  |
Correlation:0.014366
  |
Correlation:0.014366
  |
Correlation:-0.097945
  |
Correlation:0.14612
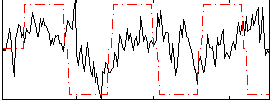  |
Component:234; Jaccard:0.071844
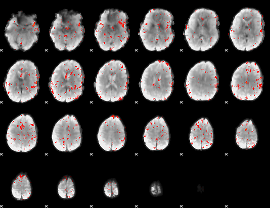 |
Component:234; Jaccard:0.076574
 |
Component:168; Jaccard:0.080878
 |
Component:156; Jaccard:0.068732
 |
Component:046; Jaccard:0.052372
 |
Component:364; Jaccard:0.030195
 |
Component:168; Jaccard:0.014513
 |
Correlation:0.1368
  |
Correlation:0.1368
  |
Correlation:0.014366
  |
Correlation:-0.13461
  |
Correlation:0.11065
  |
Correlation:0.14612
  |
Correlation:0.014366
  |
Component:313; Jaccard:0.070095
 |
Component:299; Jaccard:0.074949
 |
Component:234; Jaccard:0.074199
 |
Component:046; Jaccard:0.067996
 |
Component:364; Jaccard:0.048507
 |
Component:046; Jaccard:0.029088
 |
Component:156; Jaccard:0.013935
 |
Correlation:0.07333
  |
Correlation:0.0042478
  |
Correlation:0.1368
  |
Correlation:0.11065
  |
Correlation:0.14612
  |
Correlation:0.11065
  |
Correlation:-0.13461
  |
Component:028; Jaccard:0.067219
 |
Component:007; Jaccard:0.074862
 |
Component:364; Jaccard:0.072969
 |
Component:234; Jaccard:0.064817
 |
Component:156; Jaccard:0.048021
 |
Component:156; Jaccard:0.027907
 |
Component:046; Jaccard:0.012981
 |
Correlation:0.2315
  |
Correlation:0.28966
  |
Correlation:0.14612
  |
Correlation:0.1368
  |
Correlation:-0.13461
  |
Correlation:-0.13461
  |
Correlation:0.11065
  |
Component:364; Jaccard:0.065902
 |
Component:028; Jaccard:0.073361
 |
Component:028; Jaccard:0.07292
 |
Component:364; Jaccard:0.064514
 |
Component:234; Jaccard:0.044354
 |
Component:234; Jaccard:0.024988
 |
Component:028; Jaccard:0.012881
 |
Correlation:0.14612
  |
Correlation:0.2315
  |
Correlation:0.2315
  |
Correlation:0.14612
  |
Correlation:0.1368
  |
Correlation:0.1368
  |
Correlation:0.2315
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































