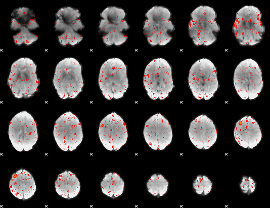
Task name: GAMBLING, TR=0.72, totally 253 volumes, 10 waves, 6 copes BACK
|
Cope: 01:PUNISH BACK
|
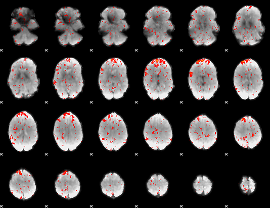
GLM result group-wise
 |
GLM binary result thresholded by 1
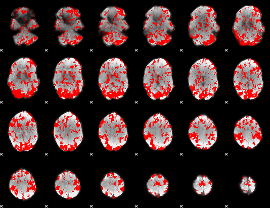 |
GLM binary result thresholded by 1.5
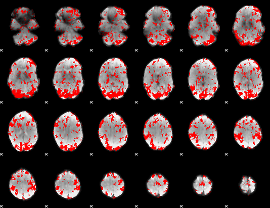 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
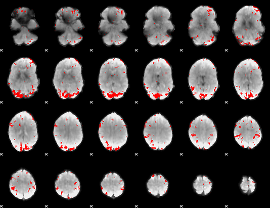 |
GLM binary result thresholded by 4
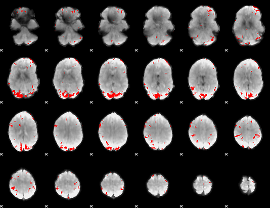 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
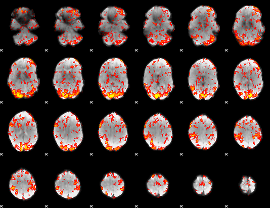 |
GLM Z-score result thresholded by 2
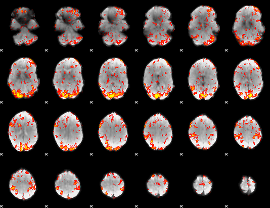 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
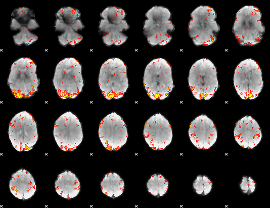 |
GLM Z-score result thresholded by 3.5
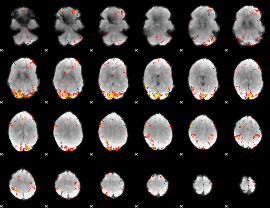 |
GLM Z-score result thresholded by 4
 |
Component:002; Jaccard:0.10893
 |
Component:002; Jaccard:0.13613
 |
Component:002; Jaccard:0.16951
 |
Component:002; Jaccard:0.20463
 |
Component:002; Jaccard:0.23484
 |
Component:002; Jaccard:0.2508
 |
Component:002; Jaccard:0.24783
 |
Correlation:0.16661
  |
Correlation:0.16661
  |
Correlation:0.16661
  |
Correlation:0.16661
  |
Correlation:0.16661
  |
Correlation:0.16661
  |
Correlation:0.16661
  |
Component:121; Jaccard:0.10367
 |
Component:243; Jaccard:0.12328
 |
Component:243; Jaccard:0.14638
 |
Component:243; Jaccard:0.16426
 |
Component:243; Jaccard:0.17804
 |
Component:243; Jaccard:0.17734
 |
Component:243; Jaccard:0.1687
 |
Correlation:0.50981
  |
Correlation:0.22744
  |
Correlation:0.22744
  |
Correlation:0.22744
  |
Correlation:0.22744
  |
Correlation:0.22744
  |
Correlation:0.22744
  |
Component:178; Jaccard:0.1025
 |
Component:121; Jaccard:0.11633
 |
Component:269; Jaccard:0.12696
 |
Component:269; Jaccard:0.14507
 |
Component:269; Jaccard:0.15557
 |
Component:269; Jaccard:0.15579
 |
Component:269; Jaccard:0.14211
 |
Correlation:0.18115
  |
Correlation:0.50981
  |
Correlation:0.09275
  |
Correlation:0.09275
  |
Correlation:0.09275
  |
Correlation:0.09275
  |
Correlation:0.09275
  |
Component:243; Jaccard:0.10051
 |
Component:178; Jaccard:0.11029
 |
Component:121; Jaccard:0.12457
 |
Component:287; Jaccard:0.12587
 |
Component:287; Jaccard:0.12742
 |
Component:287; Jaccard:0.12086
 |
Component:287; Jaccard:0.10461
 |
Correlation:0.22744
  |
Correlation:0.18115
  |
Correlation:0.50981
  |
Correlation:0.18964
  |
Correlation:0.18964
  |
Correlation:0.18964
  |
Correlation:0.18964
  |
Component:067; Jaccard:0.096885
 |
Component:067; Jaccard:0.10777
 |
Component:287; Jaccard:0.11975
 |
Component:121; Jaccard:0.12583
 |
Component:121; Jaccard:0.11983
 |
Component:121; Jaccard:0.10958
 |
Component:209; Jaccard:0.1009
 |
Correlation:0.21459
  |
Correlation:0.21459
  |
Correlation:0.18964
  |
Correlation:0.50981
  |
Correlation:0.50981
  |
Correlation:0.50981
  |
Correlation:0.15313
  |
Component:287; Jaccard:0.090884
 |
Component:269; Jaccard:0.10769
 |
Component:067; Jaccard:0.11674
 |
Component:011; Jaccard:0.11927
 |
Component:011; Jaccard:0.11914
 |
Component:209; Jaccard:0.10956
 |
Component:121; Jaccard:0.096285
 |
Correlation:0.18964
  |
Correlation:0.09275
  |
Correlation:0.21459
  |
Correlation:0.37984
  |
Correlation:0.37984
  |
Correlation:0.15313
  |
Correlation:0.50981
  |
Component:177; Jaccard:0.090494
 |
Component:287; Jaccard:0.10502
 |
Component:178; Jaccard:0.1112
 |
Component:067; Jaccard:0.11763
 |
Component:209; Jaccard:0.11262
 |
Component:011; Jaccard:0.10692
 |
Component:011; Jaccard:0.09052
 |
Correlation:0.2361
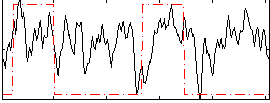  |
Correlation:0.18964
  |
Correlation:0.18115
  |
Correlation:0.21459
  |
Correlation:0.15313
  |
Correlation:0.37984
  |
Correlation:0.37984
  |
Component:203; Jaccard:0.09036
 |
Component:203; Jaccard:0.10173
 |
Component:011; Jaccard:0.11023
 |
Component:209; Jaccard:0.1104
 |
Component:067; Jaccard:0.1121
 |
Component:067; Jaccard:0.097019
 |
Component:067; Jaccard:0.081111
 |
Correlation:0.34332
  |
Correlation:0.34332
  |
Correlation:0.37984
  |
Correlation:0.15313
  |
Correlation:0.21459
  |
Correlation:0.21459
  |
Correlation:0.21459
  |
Component:269; Jaccard:0.089459
 |
Component:177; Jaccard:0.099333
 |
Component:203; Jaccard:0.10823
 |
Component:203; Jaccard:0.10844
 |
Component:203; Jaccard:0.10147
 |
Component:203; Jaccard:0.091292
 |
Component:203; Jaccard:0.077795
 |
Correlation:0.09275
  |
Correlation:0.2361
  |
Correlation:0.34332
  |
Correlation:0.34332
  |
Correlation:0.34332
  |
Correlation:0.34332
  |
Correlation:0.34332
  |
Component:033; Jaccard:0.089376
 |
Component:011; Jaccard:0.098624
 |
Component:177; Jaccard:0.10571
 |
Component:178; Jaccard:0.10641
 |
Component:383; Jaccard:0.099582
 |
Component:383; Jaccard:0.087681
 |
Component:351; Jaccard:0.069937
 |
Correlation:0.21062
 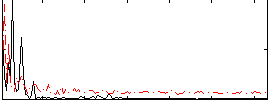 |
Correlation:0.37984
  |
Correlation:0.2361
  |
Correlation:0.18115
  |
Correlation:-0.10471
 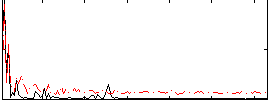 |
Correlation:-0.10471
  |
Correlation:0.31291
 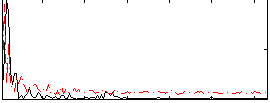 |
Cope: 02:REWARD BACK
|
GLM result group-wise
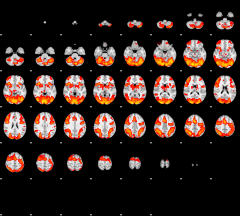 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
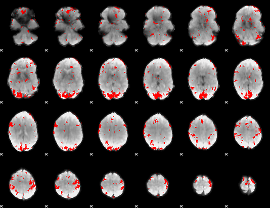 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
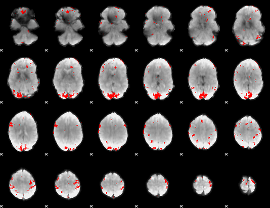 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
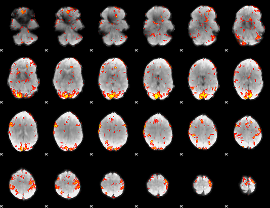 |
GLM Z-score result thresholded by 3
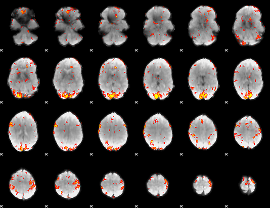 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:002; Jaccard:0.118
 |
Component:002; Jaccard:0.14964
 |
Component:002; Jaccard:0.18689
 |
Component:002; Jaccard:0.22607
 |
Component:002; Jaccard:0.26272
 |
Component:002; Jaccard:0.28919
 |
Component:002; Jaccard:0.28942
 |
Correlation:0.36946
  |
Correlation:0.36946
  |
Correlation:0.36946
  |
Correlation:0.36946
  |
Correlation:0.36946
  |
Correlation:0.36946
  |
Correlation:0.36946
  |
Component:203; Jaccard:0.096767
 |
Component:203; Jaccard:0.11098
 |
Component:243; Jaccard:0.12447
 |
Component:243; Jaccard:0.13503
 |
Component:243; Jaccard:0.13743
 |
Component:243; Jaccard:0.13521
 |
Component:243; Jaccard:0.12547
 |
Correlation:0.14196
  |
Correlation:0.14196
  |
Correlation:0.28036
  |
Correlation:0.28036
  |
Correlation:0.28036
  |
Correlation:0.28036
  |
Correlation:0.28036
  |
Component:011; Jaccard:0.094838
 |
Component:243; Jaccard:0.10887
 |
Component:203; Jaccard:0.12341
 |
Component:203; Jaccard:0.13035
 |
Component:203; Jaccard:0.12407
 |
Component:011; Jaccard:0.11218
 |
Component:011; Jaccard:0.096947
 |
Correlation:0.021594
  |
Correlation:0.28036
  |
Correlation:0.14196
  |
Correlation:0.14196
  |
Correlation:0.14196
  |
Correlation:0.021594
  |
Correlation:0.021594
  |
Component:121; Jaccard:0.093804
 |
Component:011; Jaccard:0.10703
 |
Component:011; Jaccard:0.1193
 |
Component:011; Jaccard:0.12483
 |
Component:011; Jaccard:0.12185
 |
Component:203; Jaccard:0.11157
 |
Component:269; Jaccard:0.096125
 |
Correlation:-0.085267
  |
Correlation:0.021594
  |
Correlation:0.021594
  |
Correlation:0.021594
  |
Correlation:0.021594
  |
Correlation:0.14196
  |
Correlation:0.42341
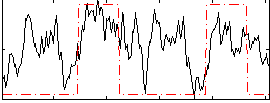  |
Component:243; Jaccard:0.092182
 |
Component:121; Jaccard:0.10149
 |
Component:121; Jaccard:0.10693
 |
Component:269; Jaccard:0.11214
 |
Component:269; Jaccard:0.11379
 |
Component:269; Jaccard:0.10883
 |
Component:203; Jaccard:0.089979
 |
Correlation:0.28036
  |
Correlation:-0.085267
  |
Correlation:-0.085267
  |
Correlation:0.42341
  |
Correlation:0.42341
  |
Correlation:0.42341
  |
Correlation:0.14196
  |
Component:300; Jaccard:0.08734
 |
Component:300; Jaccard:0.095339
 |
Component:269; Jaccard:0.10301
 |
Component:121; Jaccard:0.10556
 |
Component:185; Jaccard:0.10384
 |
Component:185; Jaccard:0.097798
 |
Component:185; Jaccard:0.088974
 |
Correlation:-0.15725
  |
Correlation:-0.15725
  |
Correlation:0.42341
  |
Correlation:-0.085267
  |
Correlation:0.42685
  |
Correlation:0.42685
  |
Correlation:0.42685
  |
Component:312; Jaccard:0.086504
 |
Component:312; Jaccard:0.092626
 |
Component:300; Jaccard:0.10049
 |
Component:185; Jaccard:0.10123
 |
Component:121; Jaccard:0.10041
 |
Component:121; Jaccard:0.091261
 |
Component:209; Jaccard:0.078954
 |
Correlation:0.25801
  |
Correlation:0.25801
  |
Correlation:-0.15725
  |
Correlation:0.42685
  |
Correlation:-0.085267
  |
Correlation:-0.085267
  |
Correlation:0.26172
 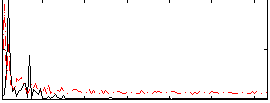 |
Component:110; Jaccard:0.082382
 |
Component:269; Jaccard:0.091931
 |
Component:185; Jaccard:0.094841
 |
Component:300; Jaccard:0.099694
 |
Component:209; Jaccard:0.095158
 |
Component:209; Jaccard:0.089736
 |
Component:121; Jaccard:0.076692
 |
Correlation:0.10993
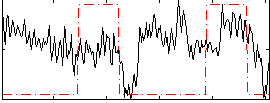  |
Correlation:0.42341
  |
Correlation:0.42685
  |
Correlation:-0.15725
  |
Correlation:0.26172
  |
Correlation:0.26172
  |
Correlation:-0.085267
  |
Component:288; Jaccard:0.082111
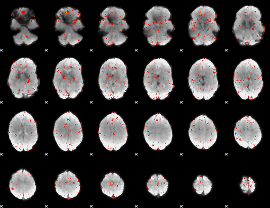 |
Component:287; Jaccard:0.088524
 |
Component:209; Jaccard:0.094711
 |
Component:209; Jaccard:0.097199
 |
Component:300; Jaccard:0.092959
 |
Component:300; Jaccard:0.081933
 |
Component:079; Jaccard:0.070051
 |
Correlation:0.29383
  |
Correlation:0.075712
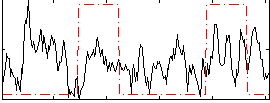  |
Correlation:0.26172
  |
Correlation:0.26172
  |
Correlation:-0.15725
  |
Correlation:-0.15725
  |
Correlation:0.27048
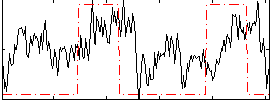  |
Component:287; Jaccard:0.082036
 |
Component:209; Jaccard:0.088398
 |
Component:312; Jaccard:0.094074
 |
Component:287; Jaccard:0.092789
 |
Component:287; Jaccard:0.087621
 |
Component:079; Jaccard:0.07823
 |
Component:300; Jaccard:0.067124
 |
Correlation:0.075712
  |
Correlation:0.26172
  |
Correlation:0.25801
  |
Correlation:0.075712
  |
Correlation:0.075712
  |
Correlation:0.27048
  |
Correlation:-0.15725
  |
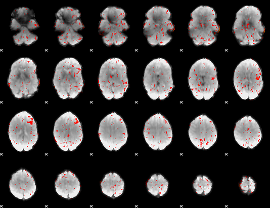
Cope: 03:PUNISH-REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
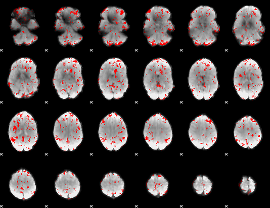 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
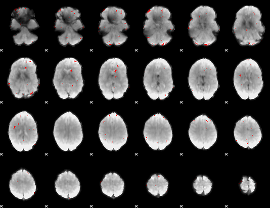 |
GLM binary result thresholded by 4
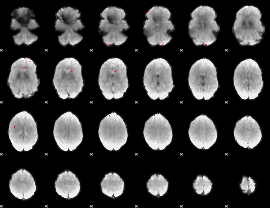 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
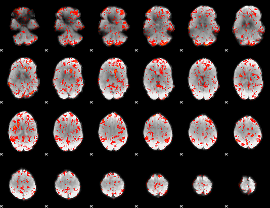 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:033; Jaccard:0.088939
 |
Component:033; Jaccard:0.097935
 |
Component:033; Jaccard:0.09674
 |
Component:033; Jaccard:0.080245
 |
Component:341; Jaccard:0.061282
 |
Component:341; Jaccard:0.040587
 |
Component:341; Jaccard:0.02017
 |
Correlation:0.073489
 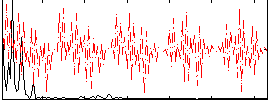 |
Correlation:0.073489
  |
Correlation:0.073489
  |
Correlation:0.073489
  |
Correlation:0.1964
  |
Correlation:0.1964
  |
Correlation:0.1964
  |
Component:290; Jaccard:0.08368
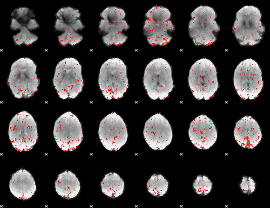 |
Component:290; Jaccard:0.091816
 |
Component:290; Jaccard:0.090354
 |
Component:341; Jaccard:0.076633
 |
Component:032; Jaccard:0.053401
 |
Component:350; Jaccard:0.031612
 |
Component:350; Jaccard:0.017469
 |
Correlation:0.18001
  |
Correlation:0.18001
  |
Correlation:0.18001
  |
Correlation:0.1964
  |
Correlation:0.27323
  |
Correlation:0.15755
  |
Correlation:0.15755
  |
Component:159; Jaccard:0.081611
 |
Component:159; Jaccard:0.083571
 |
Component:032; Jaccard:0.085538
 |
Component:290; Jaccard:0.075359
 |
Component:033; Jaccard:0.053054
 |
Component:032; Jaccard:0.031356
 |
Component:032; Jaccard:0.015894
 |
Correlation:0.1942
  |
Correlation:0.1942
  |
Correlation:0.27323
  |
Correlation:0.18001
  |
Correlation:0.073489
  |
Correlation:0.27323
  |
Correlation:0.27323
  |
Component:325; Jaccard:0.077227
 |
Component:032; Jaccard:0.082243
 |
Component:341; Jaccard:0.082405
 |
Component:032; Jaccard:0.075025
 |
Component:290; Jaccard:0.051593
 |
Component:290; Jaccard:0.02935
 |
Component:111; Jaccard:0.01503
 |
Correlation:0.13689
  |
Correlation:0.27323
  |
Correlation:0.1964
  |
Correlation:0.27323
  |
Correlation:0.18001
  |
Correlation:0.18001
  |
Correlation:0.44344
  |
Component:061; Jaccard:0.075909
 |
Component:325; Jaccard:0.080162
 |
Component:223; Jaccard:0.07765
 |
Component:223; Jaccard:0.067767
 |
Component:350; Jaccard:0.049975
 |
Component:033; Jaccard:0.028756
 |
Component:290; Jaccard:0.014808
 |
Correlation:-0.003873
  |
Correlation:0.13689
  |
Correlation:-0.14632
  |
Correlation:-0.14632
  |
Correlation:0.15755
  |
Correlation:0.073489
  |
Correlation:0.18001
  |
Component:145; Jaccard:0.074607
 |
Component:341; Jaccard:0.079647
 |
Component:350; Jaccard:0.076345
 |
Component:350; Jaccard:0.066916
 |
Component:223; Jaccard:0.048006
 |
Component:223; Jaccard:0.027733
 |
Component:033; Jaccard:0.012845
 |
Correlation:0.10915
  |
Correlation:0.1964
  |
Correlation:0.15755
  |
Correlation:0.15755
  |
Correlation:-0.14632
  |
Correlation:-0.14632
  |
Correlation:0.073489
  |
Component:032; Jaccard:0.074548
 |
Component:061; Jaccard:0.077821
 |
Component:159; Jaccard:0.074402
 |
Component:066; Jaccard:0.058689
 |
Component:348; Jaccard:0.042496
 |
Component:111; Jaccard:0.026001
 |
Component:260; Jaccard:0.012546
 |
Correlation:0.27323
  |
Correlation:-0.003873
  |
Correlation:0.1942
  |
Correlation:0.14966
  |
Correlation:0.14561
  |
Correlation:0.44344
  |
Correlation:0.18412
  |
Component:332; Jaccard:0.074239
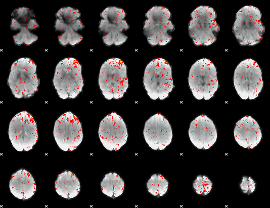 |
Component:109; Jaccard:0.07677
 |
Component:325; Jaccard:0.073672
 |
Component:325; Jaccard:0.05732
 |
Component:066; Jaccard:0.041774
 |
Component:348; Jaccard:0.02383
 |
Component:348; Jaccard:0.0125
 |
Correlation:0.054631
 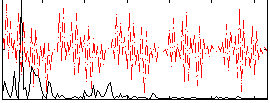 |
Correlation:0.36894
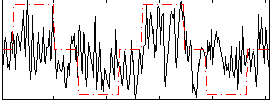 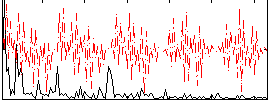 |
Correlation:0.13689
  |
Correlation:0.13689
  |
Correlation:0.14966
  |
Correlation:0.14561
  |
Correlation:0.14561
  |
Component:341; Jaccard:0.0739
 |
Component:223; Jaccard:0.075881
 |
Component:109; Jaccard:0.07262
 |
Component:348; Jaccard:0.056723
 |
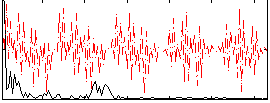
Component:252; Jaccard:0.037028
 |
Component:260; Jaccard:0.022829
 |
Component:223; Jaccard:0.012466
 |
Correlation:0.1964
  |
Correlation:-0.14632
  |
Correlation:0.36894
  |
Correlation:0.14561
  |
Correlation:0.058353
  |
Correlation:0.18412
  |
Correlation:-0.14632
  |
Component:109; Jaccard:0.073052
 |
Component:332; Jaccard:0.075436
 |
Component:332; Jaccard:0.070385
 |
Component:109; Jaccard:0.056086
 |
Component:325; Jaccard:0.036442
 |
Component:252; Jaccard:0.02189
 |
Component:358; Jaccard:0.010988
 |
Correlation:0.36894
  |
Correlation:0.054631
  |
Correlation:0.054631
  |
Correlation:0.36894
  |
Correlation:0.13689
  |
Correlation:0.058353
  |
Correlation:0.26949
  |
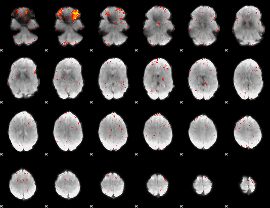
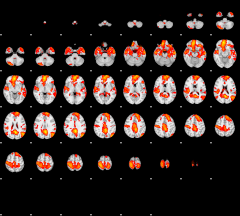
Cope: 04:neg_PUNISH BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
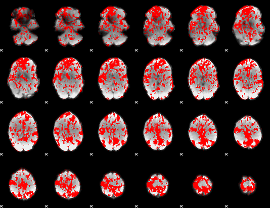 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
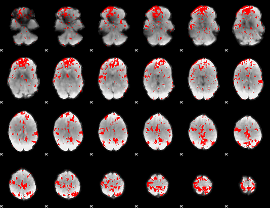 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
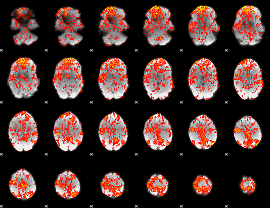 |
GLM Z-score result thresholded by 1.5
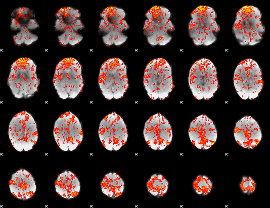 |
GLM Z-score result thresholded by 2
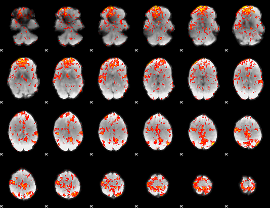 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
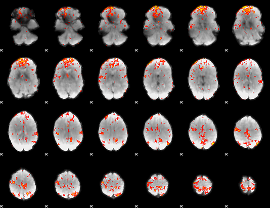 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:215; Jaccard:0.11494
 |
Component:215; Jaccard:0.1428
 |
Component:215; Jaccard:0.17418
 |
Component:215; Jaccard:0.2047
 |
Component:215; Jaccard:0.22432
 |
Component:215; Jaccard:0.22948
 |
Component:215; Jaccard:0.2206
 |
Correlation:0.13306
  |
Correlation:0.13306
  |
Correlation:0.13306
  |
Correlation:0.13306
  |
Correlation:0.13306
  |
Correlation:0.13306
  |
Correlation:0.13306
  |
Component:313; Jaccard:0.11354
 |
Component:313; Jaccard:0.13015
 |
Component:088; Jaccard:0.14299
 |
Component:088; Jaccard:0.15585
 |
Component:088; Jaccard:0.16325
 |
Component:342; Jaccard:0.15457
 |
Component:342; Jaccard:0.15383
 |
Correlation:0.025346
  |
Correlation:0.025346
  |
Correlation:0.088884
  |
Correlation:0.088884
  |
Correlation:0.088884
  |
Correlation:0.43124
  |
Correlation:0.43124
  |
Component:063; Jaccard:0.10908
 |
Component:063; Jaccard:0.12685
 |
Component:063; Jaccard:0.14238
 |
Component:063; Jaccard:0.15245
 |
Component:063; Jaccard:0.15116
 |
Component:088; Jaccard:0.15433
 |
Component:341; Jaccard:0.13563
 |
Correlation:0.021448
  |
Correlation:0.021448
  |
Correlation:0.021448
  |
Correlation:0.021448
  |
Correlation:0.021448
  |
Correlation:0.088884
  |
Correlation:0.036965
  |
Component:088; Jaccard:0.10547
 |
Component:088; Jaccard:0.12373
 |
Component:313; Jaccard:0.14175
 |
Component:313; Jaccard:0.14784
 |
Component:342; Jaccard:0.14636
 |
Component:341; Jaccard:0.13881
 |
Component:088; Jaccard:0.13213
 |
Correlation:0.088884
  |
Correlation:0.088884
  |
Correlation:0.025346
  |
Correlation:0.025346
  |
Correlation:0.43124
  |
Correlation:0.036965
  |
Correlation:0.088884
  |
Component:368; Jaccard:0.097695
 |
Component:154; Jaccard:0.10516
 |
Component:154; Jaccard:0.11828
 |
Component:342; Jaccard:0.13107
 |
Component:313; Jaccard:0.14306
 |
Component:063; Jaccard:0.13849
 |
Component:154; Jaccard:0.11691
 |
Correlation:-0.16826
  |
Correlation:0.083129
 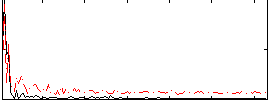 |
Correlation:0.083129
  |
Correlation:0.43124
  |
Correlation:0.025346
  |
Correlation:0.021448
  |
Correlation:0.083129
  |
Component:310; Jaccard:0.094553
 |
Component:368; Jaccard:0.10427
 |
Component:342; Jaccard:0.1115
 |
Component:154; Jaccard:0.1297
 |
Component:341; Jaccard:0.13786
 |
Component:154; Jaccard:0.12876
 |
Component:063; Jaccard:0.11588
 |
Correlation:0.11608
  |
Correlation:-0.16826
  |
Correlation:0.43124
  |
Correlation:0.083129
  |
Correlation:0.036965
  |
Correlation:0.083129
  |
Correlation:0.021448
  |
Component:154; Jaccard:0.091397
 |
Component:310; Jaccard:0.10359
 |
Component:341; Jaccard:0.11
 |
Component:341; Jaccard:0.12639
 |
Component:154; Jaccard:0.13357
 |
Component:313; Jaccard:0.12468
 |
Component:313; Jaccard:0.096199
 |
Correlation:0.083129
  |
Correlation:0.11608
  |
Correlation:0.036965
  |
Correlation:0.036965
  |
Correlation:0.083129
  |
Correlation:0.025346
  |
Correlation:0.025346
  |
Component:194; Jaccard:0.087926
 |
Component:341; Jaccard:0.094414
 |
Component:310; Jaccard:0.10838
 |
Component:310; Jaccard:0.10791
 |
Component:310; Jaccard:0.10119
 |
Component:123; Jaccard:0.094104
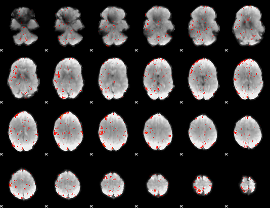 |
Component:123; Jaccard:0.08247
 |
Correlation:0.24181
 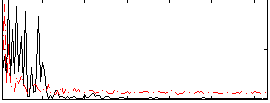 |
Correlation:0.036965
  |
Correlation:0.11608
  |
Correlation:0.11608
  |
Correlation:0.11608
  |
Correlation:0.49979
  |
Correlation:0.49979
  |
Component:322; Jaccard:0.080609
 |
Component:194; Jaccard:0.092233
 |
Component:368; Jaccard:0.10751
 |
Component:368; Jaccard:0.10174
 |
Component:123; Jaccard:0.099
 |
Component:310; Jaccard:0.086064
 |
Component:036; Jaccard:0.068919
 |
Correlation:-0.055748
 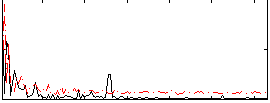 |
Correlation:0.24181
  |
Correlation:-0.16826
  |
Correlation:-0.16826
  |
Correlation:0.49979
  |
Correlation:0.11608
  |
Correlation:0.3263
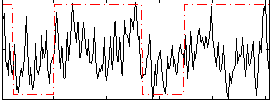 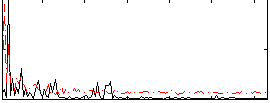 |
Component:341; Jaccard:0.079264
 |
Component:342; Jaccard:0.092094
 |
Component:123; Jaccard:0.094423
 |
Component:123; Jaccard:0.1008
 |
Component:036; Jaccard:0.091829
 |
Component:036; Jaccard:0.084604
 |
Component:310; Jaccard:0.064003
 |
Correlation:0.036965
  |
Correlation:0.43124
  |
Correlation:0.49979
  |
Correlation:0.49979
  |
Correlation:0.3263
  |
Correlation:0.3263
  |
Correlation:0.11608
  |
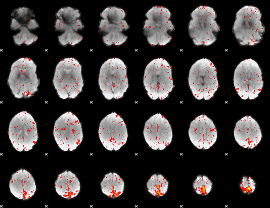
Cope: 05:neg_REWARD BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
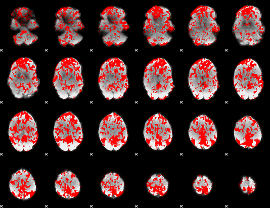 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
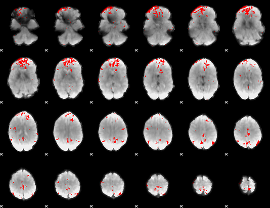 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
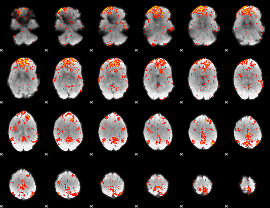 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:063; Jaccard:0.11351
 |
Component:063; Jaccard:0.13491
 |
Component:341; Jaccard:0.15749
 |
Component:341; Jaccard:0.19946
 |
Component:341; Jaccard:0.24196
 |
Component:341; Jaccard:0.27255
 |
Component:341; Jaccard:0.28062
 |
Correlation:0.14093
  |
Correlation:0.14093
  |
Correlation:0.37093
  |
Correlation:0.37093
  |
Correlation:0.37093
  |
Correlation:0.37093
  |
Correlation:0.37093
  |
Component:088; Jaccard:0.10858
 |
Component:088; Jaccard:0.13146
 |
Component:063; Jaccard:0.1563
 |
Component:088; Jaccard:0.17717
 |
Component:215; Jaccard:0.19485
 |
Component:215; Jaccard:0.20348
 |
Component:215; Jaccard:0.19354
 |
Correlation:0.20135
  |
Correlation:0.20135
  |
Correlation:0.14093
  |
Correlation:0.20135
  |
Correlation:-0.025283
  |
Correlation:-0.025283
  |
Correlation:-0.025283
  |
Component:215; Jaccard:0.10185
 |
Component:215; Jaccard:0.1232
 |
Component:088; Jaccard:0.15554
 |
Component:215; Jaccard:0.17372
 |
Component:088; Jaccard:0.1883
 |
Component:088; Jaccard:0.18966
 |
Component:342; Jaccard:0.17779
 |
Correlation:-0.025283
  |
Correlation:-0.025283
  |
Correlation:0.20135
  |
Correlation:-0.025283
  |
Correlation:0.20135
  |
Correlation:0.20135
  |
Correlation:-0.10611
  |
Component:194; Jaccard:0.10061
 |
Component:341; Jaccard:0.12138
 |
Component:215; Jaccard:0.14808
 |
Component:063; Jaccard:0.17153
 |
Component:063; Jaccard:0.17037
 |
Component:342; Jaccard:0.1762
 |
Component:088; Jaccard:0.16868
 |
Correlation:-0.14325
 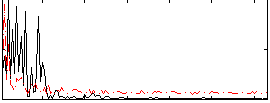 |
Correlation:0.37093
  |
Correlation:-0.025283
  |
Correlation:0.14093
  |
Correlation:0.14093
  |
Correlation:-0.10611
  |
Correlation:0.20135
  |
Component:036; Jaccard:0.094958
 |
Component:194; Jaccard:0.11157
 |
Component:036; Jaccard:0.12171
 |
Component:342; Jaccard:0.13542
 |
Component:342; Jaccard:0.15987
 |
Component:063; Jaccard:0.15847
 |
Component:063; Jaccard:0.13389
 |
Correlation:0.042308
  |
Correlation:-0.14325
  |
Correlation:0.042308
  |
Correlation:-0.10611
  |
Correlation:-0.10611
  |
Correlation:0.14093
  |
Correlation:0.14093
  |
Component:341; Jaccard:0.093801
 |
Component:036; Jaccard:0.10918
 |
Component:194; Jaccard:0.11794
 |
Component:036; Jaccard:0.12869
 |
Component:154; Jaccard:0.12638
 |
Component:154; Jaccard:0.12507
 |
Component:154; Jaccard:0.11447
 |
Correlation:0.37093
  |
Correlation:0.042308
  |
Correlation:-0.14325
  |
Correlation:0.042308
  |
Correlation:0.07893
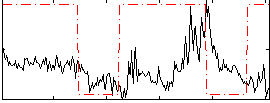 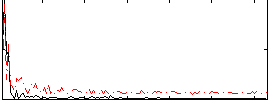 |
Correlation:0.07893
  |
Correlation:0.07893
  |
Component:313; Jaccard:0.092616
 |
Component:313; Jaccard:0.099441
 |
Component:342; Jaccard:0.11279
 |
Component:154; Jaccard:0.12054
 |
Component:036; Jaccard:0.12519
 |
Component:036; Jaccard:0.11042
 |
Component:036; Jaccard:0.089825
 |
Correlation:0.08257
 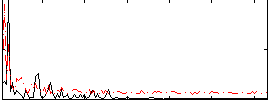 |
Correlation:0.08257
  |
Correlation:-0.10611
  |
Correlation:0.07893
  |
Correlation:0.042308
  |
Correlation:0.042308
  |
Correlation:0.042308
  |
Component:154; Jaccard:0.083471
 |
Component:154; Jaccard:0.096417
 |
Component:154; Jaccard:0.11041
 |
Component:194; Jaccard:0.1172
 |
Component:194; Jaccard:0.10396
 |
Component:194; Jaccard:0.084399
 |
Component:194; Jaccard:0.062674
 |
Correlation:0.07893
  |
Correlation:0.07893
  |
Correlation:0.07893
  |
Correlation:-0.14325
  |
Correlation:-0.14325
  |
Correlation:-0.14325
  |
Correlation:-0.14325
  |
Component:280; Jaccard:0.079966
 |
Component:342; Jaccard:0.091602
 |
Component:313; Jaccard:0.10348
 |
Component:313; Jaccard:0.099487
 |
Component:313; Jaccard:0.088205
 |
Component:313; Jaccard:0.071931
 |
Component:100; Jaccard:0.059972
 |
Correlation:-0.08766
  |
Correlation:-0.10611
  |
Correlation:0.08257
  |
Correlation:0.08257
  |
Correlation:0.08257
  |
Correlation:0.08257
  |
Correlation:0.17984
  |
Component:156; Jaccard:0.07675
 |
Component:280; Jaccard:0.083311
 |
Component:280; Jaccard:0.085084
 |
Component:145; Jaccard:0.083296
 |
Component:145; Jaccard:0.076246
 |
Component:100; Jaccard:0.06912
 |
Component:274; Jaccard:0.059478
 |
Correlation:0.042097
  |
Correlation:-0.08766
  |
Correlation:-0.08766
  |
Correlation:0.10388
  |
Correlation:0.10388
  |
Correlation:0.17984
  |
Correlation:-0.091502
 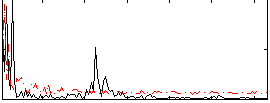 |
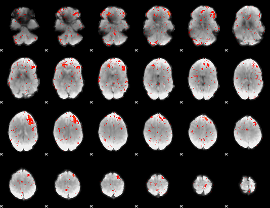
Cope: 06:REWARD-PUNISH BACK
|
GLM result group-wise
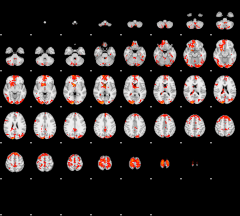 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
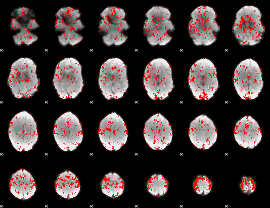 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
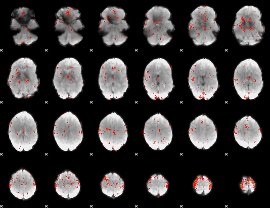 |
GLM Z-score result thresholded by 3
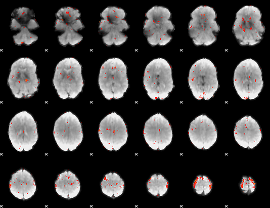 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
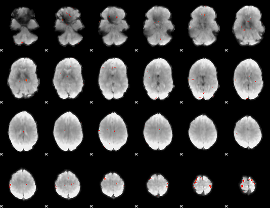 |
Component:312; Jaccard:0.097201
 |
Component:312; Jaccard:0.1105
 |
Component:312; Jaccard:0.11832
 |
Component:312; Jaccard:0.10808
 |
Component:312; Jaccard:0.080876
 |
Component:355; Jaccard:0.051991
 |
Component:355; Jaccard:0.029817
 |
Correlation:0.30328
 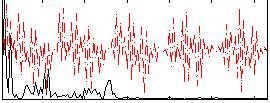 |
Correlation:0.30328
  |
Correlation:0.30328
  |
Correlation:0.30328
  |
Correlation:0.30328
  |
Correlation:0.20316
 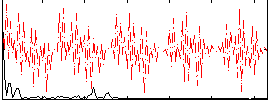 |
Correlation:0.20316
  |
Component:322; Jaccard:0.085507
 |
Component:090; Jaccard:0.089095
 |
Component:090; Jaccard:0.090951
 |
Component:355; Jaccard:0.086602
 |
Component:355; Jaccard:0.072104
 |
Component:312; Jaccard:0.050184
 |
Component:312; Jaccard:0.029258
 |
Correlation:0.0029814
  |
Correlation:0.2163
  |
Correlation:0.2163
  |
Correlation:0.20316
  |
Correlation:0.20316
  |
Correlation:0.30328
  |
Correlation:0.30328
  |
Component:090; Jaccard:0.079726
 |
Component:322; Jaccard:0.086452
 |
Component:355; Jaccard:0.090755
 |
Component:090; Jaccard:0.078491
 |
Component:090; Jaccard:0.059379
 |
Component:090; Jaccard:0.037099
 |
Component:034; Jaccard:0.019115
 |
Correlation:0.2163
  |
Correlation:0.0029814
  |
Correlation:0.20316
  |
Correlation:0.2163
  |
Correlation:0.2163
  |
Correlation:0.2163
  |
Correlation:-0.085502
  |
Component:288; Jaccard:0.079439
 |
Component:355; Jaccard:0.083322
 |
Component:288; Jaccard:0.081562
 |
Component:288; Jaccard:0.072531
 |
Component:288; Jaccard:0.053906
 |
Component:079; Jaccard:0.033175
 |
Component:246; Jaccard:0.018954
 |
Correlation:0.25474
  |
Correlation:0.20316
  |
Correlation:0.25474
  |
Correlation:0.25474
  |
Correlation:0.25474
  |
Correlation:0.22935
  |
Correlation:0.19135
  |
Component:082; Jaccard:0.076472
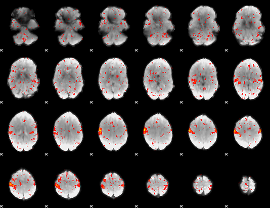 |
Component:288; Jaccard:0.082234
 |
Component:273; Jaccard:0.079172
 |
Component:273; Jaccard:0.066509
 |
Component:079; Jaccard:0.049684
 |
Component:288; Jaccard:0.032407
 |
Component:090; Jaccard:0.018913
 |
Correlation:0.21826
  |
Correlation:0.25474
  |
Correlation:0.31475
  |
Correlation:0.31475
  |
Correlation:0.22935
  |
Correlation:0.25474
  |
Correlation:0.2163
  |
Component:276; Jaccard:0.076152
 |
Component:273; Jaccard:0.082142
 |
Component:322; Jaccard:0.076852
 |
Component:246; Jaccard:0.064986
 |
Component:246; Jaccard:0.048417
 |
Component:034; Jaccard:0.032197
 |
Component:079; Jaccard:0.018896
 |
Correlation:0.13331
  |
Correlation:0.31475
  |
Correlation:0.0029814
  |
Correlation:0.19135
  |
Correlation:0.19135
  |
Correlation:-0.085502
  |
Correlation:0.22935
  |
Component:134; Jaccard:0.076067
 |
Component:082; Jaccard:0.079428
 |
Component:082; Jaccard:0.074407
 |
Component:079; Jaccard:0.061275
 |
Component:273; Jaccard:0.048073
 |
Component:246; Jaccard:0.031642
 |
Component:157; Jaccard:0.016632
 |
Correlation:-0.13338
  |
Correlation:0.21826
  |
Correlation:0.21826
  |
Correlation:0.22935
  |
Correlation:0.31475
  |
Correlation:0.19135
  |
Correlation:0.32065
  |
Component:273; Jaccard:0.075939
 |
Component:134; Jaccard:0.07695
 |
Component:110; Jaccard:0.072899
 |
Component:322; Jaccard:0.061232
 |
Component:034; Jaccard:0.045953
 |
Component:273; Jaccard:0.028329
 |
Component:028; Jaccard:0.015491
 |
Correlation:0.31475
  |
Correlation:-0.13338
  |
Correlation:-0.12368
  |
Correlation:0.0029814
  |
Correlation:-0.085502
  |
Correlation:0.31475
  |
Correlation:0.39505
  |
Component:328; Jaccard:0.075889
 |
Component:276; Jaccard:0.076883
 |
Component:276; Jaccard:0.070261
 |
Component:082; Jaccard:0.060269
 |
Component:123; Jaccard:0.04307
 |
Component:157; Jaccard:0.026822
 |
Component:216; Jaccard:0.01479
 |
Correlation:-0.22989
  |
Correlation:0.13331
  |
Correlation:0.13331
  |
Correlation:0.21826
  |
Correlation:0.44759
  |
Correlation:0.32065
  |
Correlation:0.10687
  |
Component:110; Jaccard:0.073102
 |
Component:328; Jaccard:0.075623
 |
Component:328; Jaccard:0.07023
 |
Component:110; Jaccard:0.058511
 |
Component:110; Jaccard:0.042928
 |
Component:028; Jaccard:0.026682
 |
Component:288; Jaccard:0.014344
 |
Correlation:-0.12368
  |
Correlation:-0.22989
  |
Correlation:-0.22989
  |
Correlation:-0.12368
  |
Correlation:-0.12368
  |
Correlation:0.39505
  |
Correlation:0.25474
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































