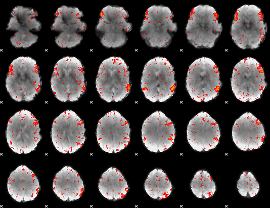
Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"FACES" BACK
|
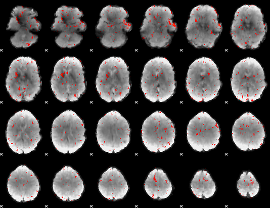
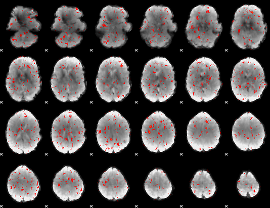
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
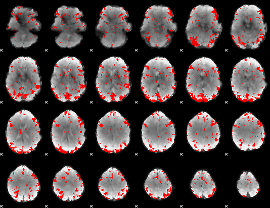 |
GLM binary result thresholded by 2.5
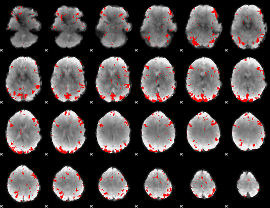 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
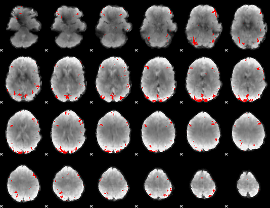 |
GLM binary result thresholded by 4
 |
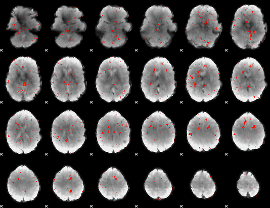
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
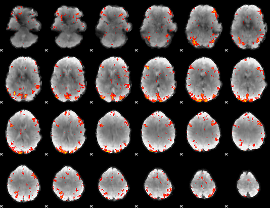 |
GLM Z-score result thresholded by 3
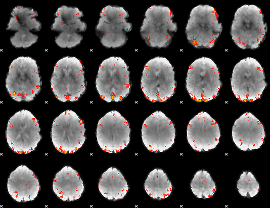 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
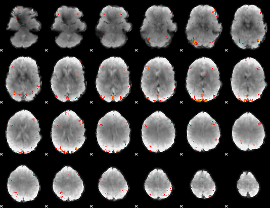 |
Component:253; Jaccard:0.13957
 |
Component:253; Jaccard:0.16946
 |
Component:030; Jaccard:0.19119
 |
Component:030; Jaccard:0.19506
 |
Component:050; Jaccard:0.18196
 |
Component:050; Jaccard:0.147
 |
Component:050; Jaccard:0.10776
 |
Correlation:0.37174
  |
Correlation:0.37174
  |
Correlation:0.63768
  |
Correlation:0.63768
  |
Correlation:0.56824
  |
Correlation:0.56824
  |
Correlation:0.56824
  |
Component:259; Jaccard:0.13523
 |
Component:030; Jaccard:0.16532
 |
Component:253; Jaccard:0.18887
 |
Component:050; Jaccard:0.19246
 |
Component:030; Jaccard:0.17007
 |
Component:029; Jaccard:0.1328
 |
Component:029; Jaccard:0.10073
 |
Correlation:0.55268
  |
Correlation:0.63768
  |
Correlation:0.37174
  |
Correlation:0.56824
  |
Correlation:0.63768
  |
Correlation:0.63244
  |
Correlation:0.63244
  |
Component:030; Jaccard:0.13079
 |
Component:050; Jaccard:0.15795
 |
Component:050; Jaccard:0.18279
 |
Component:253; Jaccard:0.18815
 |
Component:253; Jaccard:0.16559
 |
Component:253; Jaccard:0.13125
 |
Component:253; Jaccard:0.095672
 |
Correlation:0.63768
  |
Correlation:0.56824
  |
Correlation:0.56824
  |
Correlation:0.37174
  |
Correlation:0.37174
  |
Correlation:0.37174
  |
Correlation:0.37174
  |
Component:050; Jaccard:0.12448
 |
Component:259; Jaccard:0.15788
 |
Component:259; Jaccard:0.16961
 |
Component:064; Jaccard:0.16887
 |
Component:029; Jaccard:0.15649
 |
Component:030; Jaccard:0.12769
 |
Component:064; Jaccard:0.09067
 |
Correlation:0.56824
  |
Correlation:0.55268
  |
Correlation:0.55268
  |
Correlation:0.2016
  |
Correlation:0.63244
  |
Correlation:0.63768
  |
Correlation:0.2016
  |
Component:029; Jaccard:0.11599
 |
Component:029; Jaccard:0.13972
 |
Component:064; Jaccard:0.16219
 |
Component:029; Jaccard:0.16599
 |
Component:064; Jaccard:0.15557
 |
Component:064; Jaccard:0.12619
 |
Component:030; Jaccard:0.08589
 |
Correlation:0.63244
  |
Correlation:0.63244
  |
Correlation:0.2016
  |
Correlation:0.63244
  |
Correlation:0.2016
  |
Correlation:0.2016
  |
Correlation:0.63768
  |
Component:076; Jaccard:0.11161
 |
Component:064; Jaccard:0.13936
 |
Component:029; Jaccard:0.15869
 |
Component:259; Jaccard:0.15825
 |
Component:259; Jaccard:0.12745
 |
Component:259; Jaccard:0.087049
 |
Component:165; Jaccard:0.055808
 |
Correlation:0.29169
  |
Correlation:0.2016
  |
Correlation:0.63244
  |
Correlation:0.55268
  |
Correlation:0.55268
  |
Correlation:0.55268
  |
Correlation:0.42825
  |
Component:064; Jaccard:0.11017
 |
Component:076; Jaccard:0.12091
 |
Component:076; Jaccard:0.12087
 |
Component:270; Jaccard:0.10971
 |
Component:278; Jaccard:0.086914
 |
Component:165; Jaccard:0.071919
 |
Component:259; Jaccard:0.055777
 |
Correlation:0.2016
  |
Correlation:0.29169
  |
Correlation:0.29169
  |
Correlation:0.49312
 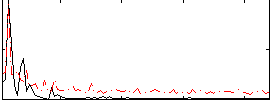 |
Correlation:0.4069
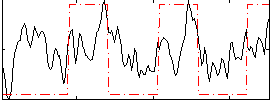  |
Correlation:0.42825
  |
Correlation:0.55268
  |
Component:204; Jaccard:0.10472
 |
Component:204; Jaccard:0.11533
 |
Component:204; Jaccard:0.11613
 |
Component:076; Jaccard:0.10668
 |
Component:270; Jaccard:0.085943
 |
Component:278; Jaccard:0.066206
 |
Component:278; Jaccard:0.044168
 |
Correlation:0.27605
  |
Correlation:0.27605
  |
Correlation:0.27605
  |
Correlation:0.29169
  |
Correlation:0.49312
  |
Correlation:0.4069
  |
Correlation:0.4069
  |
Component:278; Jaccard:0.10227
 |
Component:278; Jaccard:0.11102
 |
Component:270; Jaccard:0.11477
 |
Component:204; Jaccard:0.10453
 |
Component:165; Jaccard:0.083655
 |
Component:204; Jaccard:0.059605
 |
Component:042; Jaccard:0.039753
 |
Correlation:0.4069
  |
Correlation:0.4069
  |
Correlation:0.49312
  |
Correlation:0.27605
  |
Correlation:0.42825
  |
Correlation:0.27605
  |
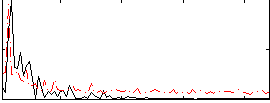
Correlation:0.48609
 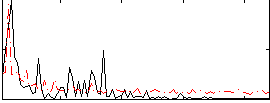 |
Component:270; Jaccard:0.096262
 |
Component:270; Jaccard:0.10956
 |
Component:278; Jaccard:0.113
 |
Component:278; Jaccard:0.10425
 |
Component:204; Jaccard:0.083373
 |
Component:042; Jaccard:0.058844
 |
Component:222; Jaccard:0.039274
 |
Correlation:0.49312
  |
Correlation:0.49312
  |
Correlation:0.4069
  |
Correlation:0.4069
  |
Correlation:0.27605
  |
Correlation:0.48609
  |
Correlation:-0.0035094
  |
Cope: 02:"SHAPES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
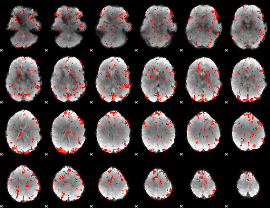 |
GLM Z-score result thresholded by 2
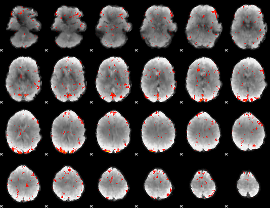 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:253; Jaccard:0.12907
 |
Component:064; Jaccard:0.13837
 |
Component:064; Jaccard:0.13838
 |
Component:064; Jaccard:0.11092
 |
Component:064; Jaccard:0.07115
 |
Component:064; Jaccard:0.03886
 |
Component:064; Jaccard:0.020088
 |
Correlation:-0.24564
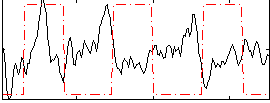  |
Correlation:0.016048
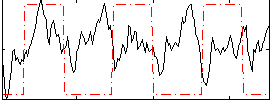  |
Correlation:0.016048
  |
Correlation:0.016048
  |
Correlation:0.016048
  |
Correlation:0.016048
  |
Correlation:0.016048
  |
Component:064; Jaccard:0.119
 |
Component:253; Jaccard:0.13835
 |
Component:253; Jaccard:0.12475
 |
Component:253; Jaccard:0.088311
 |
Component:253; Jaccard:0.052605
 |
Component:253; Jaccard:0.029158
 |
Component:253; Jaccard:0.016274
 |
Correlation:0.016048
  |
Correlation:-0.24564
  |
Correlation:-0.24564
  |
Correlation:-0.24564
  |
Correlation:-0.24564
  |
Correlation:-0.24564
  |
Correlation:-0.24564
  |
Component:204; Jaccard:0.097041
 |
Component:204; Jaccard:0.090235
 |
Component:204; Jaccard:0.068211
 |
Component:222; Jaccard:0.05213
 |
Component:222; Jaccard:0.034496
 |
Component:222; Jaccard:0.021111
 |
Component:222; Jaccard:0.011839
 |
Correlation:-0.26078
  |
Correlation:-0.26078
  |
Correlation:-0.26078
  |
Correlation:0.087105
  |
Correlation:0.087105
  |
Correlation:0.087105
  |
Correlation:0.087105
  |
Component:393; Jaccard:0.095209
 |
Component:216; Jaccard:0.085512
 |
Component:222; Jaccard:0.066804
 |
Component:050; Jaccard:0.044846
 |
Component:050; Jaccard:0.023473
 |
Component:355; Jaccard:0.014643
 |
Component:355; Jaccard:0.009524
 |
Correlation:-0.089417
 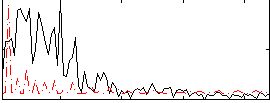 |
Correlation:-0.3145
  |
Correlation:0.087105
  |
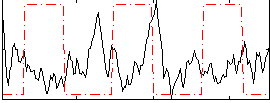
Correlation:-0.50559
  |
Correlation:-0.50559
  |
Correlation:0.013197
  |
Correlation:0.013197
  |
Component:216; Jaccard:0.091641
 |
Component:393; Jaccard:0.084967
 |
Component:050; Jaccard:0.065921
 |
Component:204; Jaccard:0.040717
 |
Component:355; Jaccard:0.023285
 |
Component:050; Jaccard:0.01124
 |
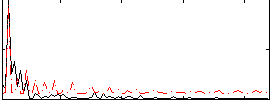
Component:007; Jaccard:0.005164
 |
Correlation:-0.3145
  |
Correlation:-0.089417
  |
Correlation:-0.50559
  |
Correlation:-0.26078
  |
Correlation:0.013197
  |
Correlation:-0.50559
  |
Correlation:-0.049969
  |
Component:076; Jaccard:0.090633
 |
Component:076; Jaccard:0.084365
 |
Component:076; Jaccard:0.064195
 |
Component:076; Jaccard:0.037855
 |
Component:027; Jaccard:0.021659
 |
Component:007; Jaccard:0.01002
 |
Component:264; Jaccard:0.005158
 |
Correlation:-0.40226
  |
Correlation:-0.40226
  |
Correlation:-0.40226
  |
Correlation:-0.40226
  |
Correlation:-0.14186
  |
Correlation:-0.049969
  |
Correlation:0.15118
  |
Component:388; Jaccard:0.084986
 |
Component:259; Jaccard:0.079914
 |
Component:259; Jaccard:0.063106
 |
Component:358; Jaccard:0.037826
 |
Component:358; Jaccard:0.019444
 |
Component:264; Jaccard:0.009564
 |
Component:050; Jaccard:0.003994
 |
Correlation:-0.22956
  |
Correlation:-0.58697
  |
Correlation:-0.58697
  |
Correlation:0.086564
  |
Correlation:0.086564
  |
Correlation:0.15118
  |
Correlation:-0.50559
  |
Component:259; Jaccard:0.084762
 |
Component:368; Jaccard:0.078816
 |
Component:216; Jaccard:0.062453
 |
Component:259; Jaccard:0.037028
 |
Component:172; Jaccard:0.019157
 |
Component:027; Jaccard:0.009084
 |
Component:207; Jaccard:0.003724
 |
Correlation:-0.58697
  |
Correlation:-0.34744
  |
Correlation:-0.3145
  |
Correlation:-0.58697
  |
Correlation:0.14689
  |
Correlation:-0.14186
  |
Correlation:0.016651
  |
Component:368; Jaccard:0.080475
 |
Component:388; Jaccard:0.076469
 |
Component:368; Jaccard:0.061409
 |
Component:216; Jaccard:0.035922
 |
Component:259; Jaccard:0.019091
 |
Component:204; Jaccard:0.007673
 |
Component:089; Jaccard:0.003513
 |
Correlation:-0.34744
  |
Correlation:-0.22956
  |
Correlation:-0.34744
  |
Correlation:-0.3145
  |
Correlation:-0.58697
  |
Correlation:-0.26078
  |
Correlation:0.10057
  |
Component:190; Jaccard:0.078897
 |
Component:050; Jaccard:0.074948
 |
Component:393; Jaccard:0.057668
 |
Component:027; Jaccard:0.035168
 |
Component:204; Jaccard:0.01858
 |
Component:358; Jaccard:0.00761
 |
Component:223; Jaccard:0.003267
 |
Correlation:-0.33668
  |
Correlation:-0.50559
  |
Correlation:-0.089417
  |
Correlation:-0.14186
  |
Correlation:-0.26078
  |
Correlation:0.086564
  |
Correlation:0.1353
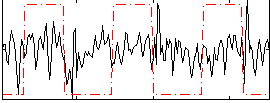 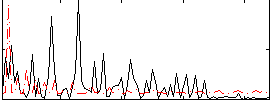 |
Cope: 03:"FACES-SHAPES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
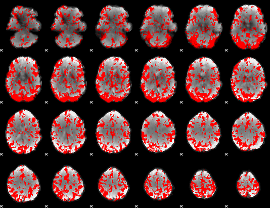 |
GLM binary result thresholded by 2
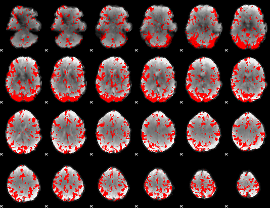 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:278; Jaccard:0.11236
 |
Component:029; Jaccard:0.14876
 |
Component:029; Jaccard:0.19883
 |
Component:029; Jaccard:0.25763
 |
Component:029; Jaccard:0.31334
 |
Component:029; Jaccard:0.34643
 |
Component:029; Jaccard:0.34558
 |
Correlation:0.37368
  |
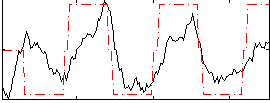
Correlation:0.60925
 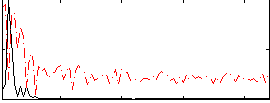 |
Correlation:0.60925
  |
Correlation:0.60925
  |
Correlation:0.60925
  |
Correlation:0.60925
  |
Correlation:0.60925
  |
Component:029; Jaccard:0.11166
 |
Component:278; Jaccard:0.13499
 |
Component:118; Jaccard:0.16509
 |
Component:030; Jaccard:0.20001
 |
Component:030; Jaccard:0.22406
 |
Component:030; Jaccard:0.23232
 |
Component:030; Jaccard:0.21656
 |
Correlation:0.60925
  |
Correlation:0.37368
  |
Correlation:0.65971
 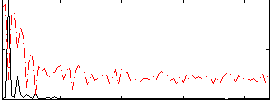 |
Correlation:0.60779
  |
Correlation:0.60779
  |
Correlation:0.60779
  |
Correlation:0.60779
  |
Component:118; Jaccard:0.1068
 |
Component:118; Jaccard:0.1348
 |
Component:030; Jaccard:0.16498
 |
Component:118; Jaccard:0.19201
 |
Component:118; Jaccard:0.20935
 |
Component:118; Jaccard:0.21226
 |
Component:118; Jaccard:0.20269
 |
Correlation:0.65971
  |
Correlation:0.65971
  |
Correlation:0.60779
  |
Correlation:0.65971
  |
Correlation:0.65971
  |
Correlation:0.65971
  |
Correlation:0.65971
  |
Component:030; Jaccard:0.10165
 |
Component:030; Jaccard:0.13134
 |
Component:278; Jaccard:0.15761
 |
Component:278; Jaccard:0.1718
 |
Component:342; Jaccard:0.17864
 |
Component:342; Jaccard:0.17632
 |
Component:050; Jaccard:0.15951
 |
Correlation:0.60779
  |
Correlation:0.60779
  |
Correlation:0.37368
  |
Correlation:0.37368
  |
Correlation:0.68268
  |
Correlation:0.68268
  |
Correlation:0.58246
 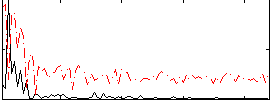 |
Component:342; Jaccard:0.099952
 |
Component:342; Jaccard:0.12269
 |
Component:342; Jaccard:0.14749
 |
Component:342; Jaccard:0.16879
 |
Component:278; Jaccard:0.17437
 |
Component:050; Jaccard:0.17277
 |
Component:342; Jaccard:0.15642
 |
Correlation:0.68268
  |
Correlation:0.68268
  |
Correlation:0.68268
  |
Correlation:0.68268
  |
Correlation:0.37368
  |
Correlation:0.58246
  |
Correlation:0.68268
  |
Component:259; Jaccard:0.099085
 |
Component:259; Jaccard:0.1136
 |
Component:050; Jaccard:0.13654
 |
Component:050; Jaccard:0.15783
 |
Component:050; Jaccard:0.17019
 |
Component:278; Jaccard:0.15817
 |
Component:278; Jaccard:0.13367
 |
Correlation:0.61816
  |
Correlation:0.61816
  |
Correlation:0.58246
  |
Correlation:0.58246
  |
Correlation:0.58246
  |
Correlation:0.37368
  |
Correlation:0.37368
  |
Component:138; Jaccard:0.096322
 |
Component:050; Jaccard:0.11262
 |
Component:259; Jaccard:0.12614
 |
Component:259; Jaccard:0.13184
 |
Component:259; Jaccard:0.12973
 |
Component:259; Jaccard:0.11706
 |
Component:165; Jaccard:0.10717
 |
Correlation:0.42927
 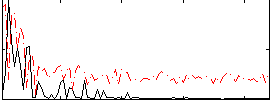 |
Correlation:0.58246
  |
Correlation:0.61816
  |
Correlation:0.61816
  |
Correlation:0.61816
  |
Correlation:0.61816
  |
Correlation:0.43535
  |
Component:394; Jaccard:0.093646
 |
Component:138; Jaccard:0.1077
 |
Component:138; Jaccard:0.11719
 |
Component:138; Jaccard:0.11937
 |
Component:165; Jaccard:0.11183
 |
Component:165; Jaccard:0.11144
 |
Component:259; Jaccard:0.099361
 |
Correlation:0.2659
  |
Correlation:0.42927
  |
Correlation:0.42927
  |
Correlation:0.42927
  |
Correlation:0.43535
  |
Correlation:0.43535
  |
Correlation:0.61816
  |
Component:050; Jaccard:0.091538
 |
Component:394; Jaccard:0.10329
 |
Component:394; Jaccard:0.11145
 |
Component:394; Jaccard:0.11477
 |
Component:138; Jaccard:0.11132
 |
Component:394; Jaccard:0.10099
 |
Component:394; Jaccard:0.089153
 |
Correlation:0.58246
  |
Correlation:0.2659
  |
Correlation:0.2659
  |
Correlation:0.2659
  |
Correlation:0.42927
  |
Correlation:0.2659
  |
Correlation:0.2659
  |
Component:265; Jaccard:0.089307
 |
Component:168; Jaccard:0.097712
 |
Component:168; Jaccard:0.10791
 |
Component:168; Jaccard:0.11302
 |
Component:168; Jaccard:0.11048
 |
Component:168; Jaccard:0.10002
 |
Component:341; Jaccard:0.084306
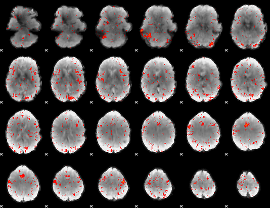 |
Correlation:0.34065
  |
Correlation:0.4111
  |
Correlation:0.4111
  |
Correlation:0.4111
  |
Correlation:0.4111
  |
Correlation:0.4111
  |
Correlation:0.50732
  |
Cope: 04:"neg_FACES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
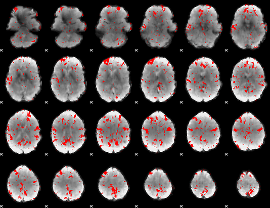 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
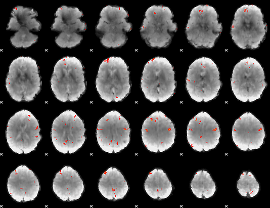 |
GLM Z-score result thresholded by 4
 |
Component:324; Jaccard:0.12342
 |
Component:324; Jaccard:0.15567
 |
Component:324; Jaccard:0.17709
 |
Component:324; Jaccard:0.16485
 |
Component:324; Jaccard:0.12475
 |
Component:324; Jaccard:0.073136
 |
Component:324; Jaccard:0.038754
 |
Correlation:0.38741
  |
Correlation:0.38741
  |
Correlation:0.38741
  |
Correlation:0.38741
  |
Correlation:0.38741
  |
Correlation:0.38741
  |
Correlation:0.38741
  |
Component:384; Jaccard:0.11661
 |
Component:384; Jaccard:0.12886
 |
Component:384; Jaccard:0.1302
 |
Component:384; Jaccard:0.10946
 |
Component:384; Jaccard:0.075883
 |
Component:384; Jaccard:0.041807
 |
Component:384; Jaccard:0.022912
 |
Correlation:0.091972
  |
Correlation:0.091972
  |
Correlation:0.091972
  |
Correlation:0.091972
  |
Correlation:0.091972
  |
Correlation:0.091972
  |
Correlation:0.091972
  |
Component:295; Jaccard:0.1053
 |
Component:295; Jaccard:0.11699
 |
Component:295; Jaccard:0.11673
 |
Component:295; Jaccard:0.095955
 |
Component:295; Jaccard:0.065487
 |
Component:142; Jaccard:0.039969
 |
Component:142; Jaccard:0.022233
 |
Correlation:0.055546
  |
Correlation:0.055546
  |
Correlation:0.055546
  |
Correlation:0.055546
  |
Correlation:0.055546
  |
Correlation:0.36123
  |
Correlation:0.36123
  |
Component:102; Jaccard:0.094939
 |
Component:102; Jaccard:0.10326
 |
Component:142; Jaccard:0.10416
 |
Component:142; Jaccard:0.088613
 |
Component:142; Jaccard:0.063498
 |
Component:295; Jaccard:0.039816
 |
Component:295; Jaccard:0.022037
 |
Correlation:-0.088336
  |
Correlation:-0.088336
  |
Correlation:0.36123
  |
Correlation:0.36123
  |
Correlation:0.36123
  |
Correlation:0.055546
  |
Correlation:0.055546
  |
Component:346; Jaccard:0.094161
 |
Component:142; Jaccard:0.10325
 |
Component:102; Jaccard:0.10147
 |
Component:102; Jaccard:0.085547
 |
Component:102; Jaccard:0.059636
 |
Component:102; Jaccard:0.035288
 |
Component:102; Jaccard:0.02025
 |
Correlation:0.11904
  |
Correlation:0.36123
  |
Correlation:-0.088336
  |
Correlation:-0.088336
  |
Correlation:-0.088336
  |
Correlation:-0.088336
  |
Correlation:-0.088336
  |
Component:093; Jaccard:0.091991
 |
Component:346; Jaccard:0.10088
 |
Component:346; Jaccard:0.09887
 |
Component:346; Jaccard:0.081097
 |
Component:346; Jaccard:0.056007
 |
Component:346; Jaccard:0.032897
 |
Component:346; Jaccard:0.01705
 |
Correlation:-0.3663
  |
Correlation:0.11904
  |
Correlation:0.11904
  |
Correlation:0.11904
  |
Correlation:0.11904
  |
Correlation:0.11904
  |
Correlation:0.11904
  |
Component:142; Jaccard:0.0912
 |
Component:093; Jaccard:0.093572
 |
Component:093; Jaccard:0.087832
 |
Component:077; Jaccard:0.071276
 |
Component:093; Jaccard:0.049096
 |
Component:093; Jaccard:0.030592
 |
Component:093; Jaccard:0.016875
 |
Correlation:0.36123
  |
Correlation:-0.3663
  |
Correlation:-0.3663
  |
Correlation:0.050216
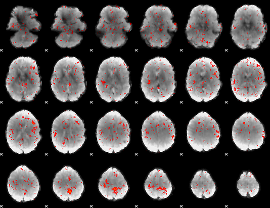
  |
Correlation:-0.3663
  |
Correlation:-0.3663
  |
Correlation:-0.3663
  |
Component:037; Jaccard:0.085637
 |
Component:077; Jaccard:0.088512
 |
Component:077; Jaccard:0.08547
 |
Component:093; Jaccard:0.07
 |
Component:077; Jaccard:0.044913
 |
Component:077; Jaccard:0.02547
 |
Component:331; Jaccard:0.013376
 |
Correlation:-0.16834
  |
Correlation:0.050216
  |
Correlation:0.050216
  |
Correlation:-0.3663
  |
Correlation:0.050216
  |
Correlation:0.050216
  |
Correlation:0.16896
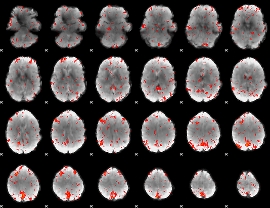
  |
Component:077; Jaccard:0.083236
 |
Component:262; Jaccard:0.086354
 |
Component:262; Jaccard:0.082762
 |
Component:262; Jaccard:0.067426
 |
Component:262; Jaccard:0.043965
 |
Component:243; Jaccard:0.023569
 |
Component:073; Jaccard:0.012469
 |
Correlation:0.050216
  |
Correlation:0.11903
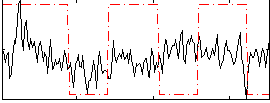  |
Correlation:0.11903
  |
Correlation:0.11903
  |
Correlation:0.11903
  |
Correlation:-0.01416
  |
Correlation:0.31115
 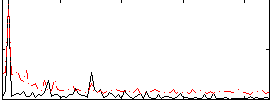 |
Component:242; Jaccard:0.079384
 |
Component:037; Jaccard:0.086274
 |
Component:037; Jaccard:0.077297
 |
Component:237; Jaccard:0.061702
 |
Component:243; Jaccard:0.040552
 |
Component:331; Jaccard:0.022985
 |
Component:243; Jaccard:0.01219
 |
Correlation:-0.19196
  |
Correlation:-0.16834
  |
Correlation:-0.16834
  |
Correlation:0.29494
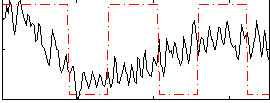  |
Correlation:-0.01416
  |
Correlation:0.16896
  |
Correlation:-0.01416
  |
Cope: 05:"neg_SHAPES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
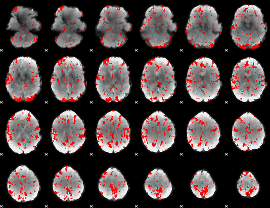 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
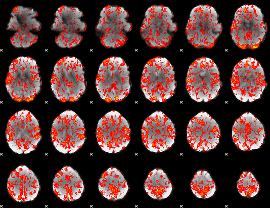
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
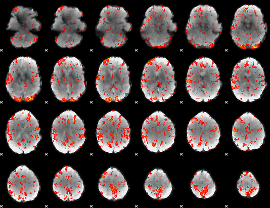
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:118; Jaccard:0.11389
 |
Component:118; Jaccard:0.14435
 |
Component:118; Jaccard:0.16738
 |
Component:118; Jaccard:0.17198
 |
Component:118; Jaccard:0.15103
 |
Component:118; Jaccard:0.10954
 |
Component:118; Jaccard:0.074044
 |
Correlation:0.58432
  |
Correlation:0.58432
  |
Correlation:0.58432
  |
Correlation:0.58432
  |
Correlation:0.58432
  |
Correlation:0.58432
  |
Correlation:0.58432
  |
Component:166; Jaccard:0.10454
 |
Component:166; Jaccard:0.11975
 |
Component:166; Jaccard:0.12952
 |
Component:166; Jaccard:0.12413
 |
Component:166; Jaccard:0.098953
 |
Component:384; Jaccard:0.068072
 |
Component:058; Jaccard:0.040572
 |
Correlation:0.52991
  |
Correlation:0.52991
  |
Correlation:0.52991
  |
Correlation:0.52991
  |
Correlation:0.52991
  |
Correlation:0.0047841
  |
Correlation:0.18228
  |
Component:037; Jaccard:0.10192
 |
Component:384; Jaccard:0.11664
 |
Component:384; Jaccard:0.12493
 |
Component:384; Jaccard:0.12104
 |
Component:384; Jaccard:0.097981
 |
Component:166; Jaccard:0.06595
 |
Component:166; Jaccard:0.040081
 |
Correlation:0.18516
  |
Correlation:0.0047841
  |
Correlation:0.0047841
  |
Correlation:0.0047841
  |
Correlation:0.0047841
  |
Correlation:0.52991
  |
Correlation:0.52991
  |
Component:384; Jaccard:0.10064
 |
Component:037; Jaccard:0.11594
 |
Component:037; Jaccard:0.12015
 |
Component:037; Jaccard:0.11233
 |
Component:037; Jaccard:0.090492
 |
Component:058; Jaccard:0.065002
 |
Component:384; Jaccard:0.039764
 |
Correlation:0.0047841
  |
Correlation:0.18516
  |
Correlation:0.18516
  |
Correlation:0.18516
  |
Correlation:0.18516
  |
Correlation:0.18228
  |
Correlation:0.0047841
  |
Component:025; Jaccard:0.09607
 |
Component:295; Jaccard:0.10648
 |
Component:295; Jaccard:0.11624
 |
Component:295; Jaccard:0.10715
 |
Component:058; Jaccard:0.08935
 |
Component:102; Jaccard:0.06138
 |
Component:102; Jaccard:0.036142
 |
Correlation:0.42611
  |
Correlation:0.11084
  |
Correlation:0.11084
  |
Correlation:0.11084
  |
Correlation:0.18228
  |
Correlation:0.14219
  |
Correlation:0.14219
  |
Component:138; Jaccard:0.09252
 |
Component:025; Jaccard:0.106
 |
Component:058; Jaccard:0.10898
 |
Component:102; Jaccard:0.10703
 |
Component:102; Jaccard:0.087596
 |
Component:037; Jaccard:0.059198
 |
Component:295; Jaccard:0.035322
 |
Correlation:0.38724
  |
Correlation:0.42611
  |
Correlation:0.18228
  |
Correlation:0.14219
  |
Correlation:0.14219
  |
Correlation:0.18516
  |
Correlation:0.11084
  |
Component:295; Jaccard:0.091599
 |
Component:138; Jaccard:0.10041
 |
Component:025; Jaccard:0.10891
 |
Component:058; Jaccard:0.10591
 |
Component:295; Jaccard:0.085764
 |
Component:295; Jaccard:0.058418
 |
Component:077; Jaccard:0.034314
 |
Correlation:0.11084
  |
Correlation:0.38724
  |
Correlation:0.42611
  |
Correlation:0.18228
  |
Correlation:0.11084
  |
Correlation:0.11084
  |
Correlation:-0.068178
  |
Component:102; Jaccard:0.08716
 |
Component:102; Jaccard:0.1004
 |
Component:102; Jaccard:0.1084
 |
Component:025; Jaccard:0.099646
 |
Component:025; Jaccard:0.079557
 |
Component:025; Jaccard:0.055102
 |
Component:025; Jaccard:0.033659
 |
Correlation:0.14219
  |
Correlation:0.14219
  |
Correlation:0.14219
  |
Correlation:0.42611
  |
Correlation:0.42611
  |
Correlation:0.42611
  |
Correlation:0.42611
  |
Component:093; Jaccard:0.08688
 |
Component:058; Jaccard:0.099387
 |
Component:245; Jaccard:0.10214
 |
Component:245; Jaccard:0.091212
 |
Component:077; Jaccard:0.075158
 |
Component:077; Jaccard:0.054535
 |
Component:037; Jaccard:0.033515
 |
Correlation:0.45556
  |
Correlation:0.18228
  |
Correlation:-0.059544
  |
Correlation:-0.059544
  |
Correlation:-0.068178
  |
Correlation:-0.068178
  |
Correlation:0.18516
  |
Component:058; Jaccard:0.086762
 |
Component:245; Jaccard:0.09721
 |
Component:138; Jaccard:0.10018
 |
Component:077; Jaccard:0.091095
 |
Component:093; Jaccard:0.070694
 |
Component:093; Jaccard:0.048611
 |
Component:093; Jaccard:0.030499
 |
Correlation:0.18228
  |
Correlation:-0.059544
  |
Correlation:0.38724
  |
Correlation:-0.068178
  |
Correlation:0.45556
  |
Correlation:0.45556
  |
Correlation:0.45556
  |
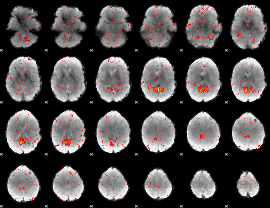
Cope: 06:"SHAPES-FACES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
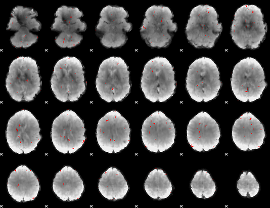 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:324; Jaccard:0.09382
 |
Component:324; Jaccard:0.093417
 |
Component:075; Jaccard:0.079417
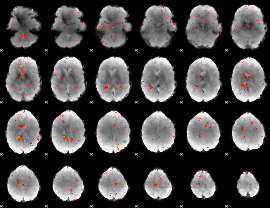 |
Component:075; Jaccard:0.058716
 |
Component:075; Jaccard:0.03368
 |
Component:073; Jaccard:0.012972
 |
Component:065; Jaccard:0.007051
 |
Correlation:0.32831
  |
Correlation:0.32831
  |
Correlation:0.51175
  |
Correlation:0.51175
  |
Correlation:0.51175
  |
Correlation:0.42664
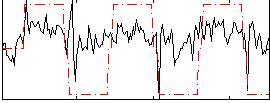  |
Correlation:0.33954
 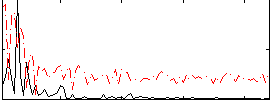 |
Component:285; Jaccard:0.090724
 |
Component:285; Jaccard:0.087288
 |
Component:324; Jaccard:0.075917
 |
Component:285; Jaccard:0.048307
 |
Component:073; Jaccard:0.02624
 |
Component:075; Jaccard:0.012089
 |
Component:073; Jaccard:0.005867
 |
Correlation:0.19721
  |
Correlation:0.19721
  |
Correlation:0.32831
  |
Correlation:0.19721
  |
Correlation:0.42664
  |
Correlation:0.51175
  |
Correlation:0.42664
  |
Component:010; Jaccard:0.082474
 |
Component:075; Jaccard:0.076818
 |
Component:285; Jaccard:0.071404
 |
Component:324; Jaccard:0.046489
 |
Component:285; Jaccard:0.025206
 |
Component:285; Jaccard:0.008503
 |
Component:075; Jaccard:0.005828
 |
Correlation:0.15546
  |
Correlation:0.51175
  |
Correlation:0.19721
  |
Correlation:0.32831
  |
Correlation:0.19721
  |
Correlation:0.19721
  |
Correlation:0.51175
  |
Component:382; Jaccard:0.072556
 |
Component:010; Jaccard:0.076252
 |
Component:073; Jaccard:0.061887
 |
Component:073; Jaccard:0.045469
 |
Component:324; Jaccard:0.022864
 |
Component:065; Jaccard:0.007688
 |
Component:313; Jaccard:0.003201
 |
Correlation:-0.11799
  |
Correlation:0.15546
  |
Correlation:0.42664
  |
Correlation:0.42664
  |
Correlation:0.32831
  |
Correlation:0.33954
  |
Correlation:-0.01745
  |
Component:161; Jaccard:0.072421
 |
Component:161; Jaccard:0.070823
 |
Component:010; Jaccard:0.060894
 |
Component:010; Jaccard:0.03708
 |
Component:100; Jaccard:0.015878
 |
Component:324; Jaccard:0.007487
 |
Component:182; Jaccard:0.002517
 |
Correlation:0.27152
  |
Correlation:0.27152
  |
Correlation:0.15546
  |
Correlation:0.15546
  |
Correlation:0.33656
  |
Correlation:0.32831
  |
Correlation:0.067542
  |
Component:333; Jaccard:0.072241
 |
Component:382; Jaccard:0.070798
 |
Component:188; Jaccard:0.057815
 |
Component:188; Jaccard:0.035023
 |
Component:010; Jaccard:0.015823
 |
Component:269; Jaccard:0.006539
 |
Component:285; Jaccard:0.002464
 |
Correlation:0.11652
  |
Correlation:-0.11799
  |
Correlation:0.17034
  |
Correlation:0.17034
  |
Correlation:0.15546
  |
Correlation:0.063081
  |
Correlation:0.19721
  |
Component:142; Jaccard:0.070904
 |
Component:188; Jaccard:0.06737
 |
Component:161; Jaccard:0.055999
 |
Component:311; Jaccard:0.033336
 |
Component:237; Jaccard:0.015595
 |
Component:182; Jaccard:0.006166
 |
Component:172; Jaccard:0.002429
 |
Correlation:0.26469
  |
Correlation:0.17034
  |
Correlation:0.27152
  |
Correlation:0.25735
  |
Correlation:0.17936
  |
Correlation:0.067542
  |
Correlation:0.17369
  |
Component:311; Jaccard:0.069895
 |
Component:311; Jaccard:0.066637
 |
Component:382; Jaccard:0.054953
 |
Component:161; Jaccard:0.033221
 |
Component:161; Jaccard:0.015392
 |
Component:100; Jaccard:0.005825
 |
Component:389; Jaccard:0.002333
 |
Correlation:0.25735
  |
Correlation:0.25735
  |
Correlation:-0.11799
  |
Correlation:0.27152
  |
Correlation:0.27152
  |
Correlation:0.33656
  |
Correlation:0.26283
  |
Component:075; Jaccard:0.068148
 |
Component:142; Jaccard:0.065556
 |
Component:311; Jaccard:0.053676
 |
Component:237; Jaccard:0.032888
 |
Component:311; Jaccard:0.014953
 |
Component:311; Jaccard:0.005584
 |
Component:100; Jaccard:0.002324
 |
Correlation:0.51175
  |
Correlation:0.26469
  |
Correlation:0.25735
  |
Correlation:0.17936
  |
Correlation:0.25735
  |
Correlation:0.25735
  |
Correlation:0.33656
  |
Component:188; Jaccard:0.067644
 |
Component:073; Jaccard:0.065489
 |
Component:237; Jaccard:0.051578
 |
Component:389; Jaccard:0.030805
 |
Component:333; Jaccard:0.014631
 |
Component:010; Jaccard:0.005356
 |
Component:151; Jaccard:0.002269
 |
Correlation:0.17034
  |
Correlation:0.42664
  |
Correlation:0.17936
  |
Correlation:0.26283
  |
Correlation:0.11652
  |
Correlation:0.15546
  |
Correlation:0.23026
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































