Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:070; Jaccard:0.16464
 |
Component:343; Jaccard:0.19839
 |
Component:343; Jaccard:0.25088
 |
Component:343; Jaccard:0.30555
 |
Component:343; Jaccard:0.35678
 |
Component:343; Jaccard:0.39984
 |
Component:343; Jaccard:0.43139
 |
Correlation:0.12146
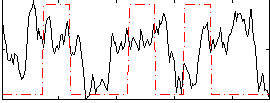 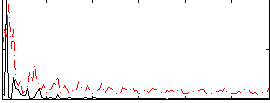 |
Correlation:0.1632
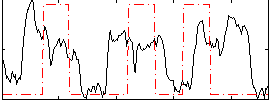 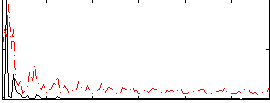 |
Correlation:0.1632
  |
Correlation:0.1632
  |
Correlation:0.1632
  |
Correlation:0.1632
  |
Correlation:0.1632
  |
Component:343; Jaccard:0.15422
 |
Component:070; Jaccard:0.19635
 |
Component:070; Jaccard:0.22636
 |
Component:153; Jaccard:0.26288
 |
Component:153; Jaccard:0.28971
 |
Component:153; Jaccard:0.31015
 |
Component:153; Jaccard:0.32081
 |
Correlation:0.1632
  |
Correlation:0.12146
  |
Correlation:0.12146
  |
Correlation:0.19099
  |
Correlation:0.19099
  |
Correlation:0.19099
  |
Correlation:0.19099
  |
Component:375; Jaccard:0.15156
 |
Component:375; Jaccard:0.18951
 |
Component:375; Jaccard:0.2257
 |
Component:375; Jaccard:0.25532
 |
Component:375; Jaccard:0.27037
 |
Component:375; Jaccard:0.27361
 |
Component:375; Jaccard:0.26397
 |
Correlation:0.20518
  |
Correlation:0.20518
  |
Correlation:0.20518
  |
Correlation:0.20518
  |
Correlation:0.20518
  |
Correlation:0.20518
  |
Correlation:0.20518
  |
Component:153; Jaccard:0.14691
 |
Component:153; Jaccard:0.18476
 |
Component:153; Jaccard:0.22485
 |
Component:070; Jaccard:0.24596
 |
Component:070; Jaccard:0.2567
 |
Component:371; Jaccard:0.26031
 |
Component:371; Jaccard:0.26154
 |
Correlation:0.19099
  |
Correlation:0.19099
  |
Correlation:0.19099
  |
Correlation:0.12146
  |
Correlation:0.12146
  |
Correlation:0.084793
  |
Correlation:0.084793
  |
Component:250; Jaccard:0.14177
 |
Component:371; Jaccard:0.17085
 |
Component:371; Jaccard:0.20439
 |
Component:371; Jaccard:0.23234
 |
Component:371; Jaccard:0.25135
 |
Component:070; Jaccard:0.25473
 |
Component:070; Jaccard:0.24329
 |
Correlation:0.067939
  |
Correlation:0.084793
  |
Correlation:0.084793
  |
Correlation:0.084793
  |
Correlation:0.084793
  |
Correlation:0.12146
  |
Correlation:0.12146
  |
Component:371; Jaccard:0.13959
 |
Component:250; Jaccard:0.16083
 |
Component:225; Jaccard:0.18267
 |
Component:225; Jaccard:0.20588
 |
Component:225; Jaccard:0.22328
 |
Component:225; Jaccard:0.23438
 |
Component:225; Jaccard:0.23913
 |
Correlation:0.084793
  |
Correlation:0.067939
  |
Correlation:0.15447
  |
Correlation:0.15447
  |
Correlation:0.15447
  |
Correlation:0.15447
  |
Correlation:0.15447
  |
Component:225; Jaccard:0.12801
 |
Component:225; Jaccard:0.15496
 |
Component:250; Jaccard:0.17686
 |
Component:178; Jaccard:0.20149
 |
Component:178; Jaccard:0.2197
 |
Component:178; Jaccard:0.23188
 |
Component:178; Jaccard:0.2362
 |
Correlation:0.15447
  |
Correlation:0.15447
  |
Correlation:0.067939
  |
Correlation:0.10494
  |
Correlation:0.10494
  |
Correlation:0.10494
  |
Correlation:0.10494
  |
Component:178; Jaccard:0.1217
 |
Component:178; Jaccard:0.14909
 |
Component:178; Jaccard:0.17613
 |
Component:250; Jaccard:0.18444
 |
Component:250; Jaccard:0.18684
 |
Component:250; Jaccard:0.18244
 |
Component:250; Jaccard:0.173
 |
Correlation:0.10494
  |
Correlation:0.10494
  |
Correlation:0.10494
  |
Correlation:0.067939
  |
Correlation:0.067939
  |
Correlation:0.067939
  |
Correlation:0.067939
  |
Component:347; Jaccard:0.12075
 |
Component:334; Jaccard:0.13735
 |
Component:303; Jaccard:0.15111
 |
Component:303; Jaccard:0.16431
 |
Component:303; Jaccard:0.17026
 |
Component:303; Jaccard:0.17113
 |
Component:303; Jaccard:0.1641
 |
Correlation:0.28488
  |
Correlation:0.23761
  |
Correlation:0.32548
  |
Correlation:0.32548
  |
Correlation:0.32548
  |
Correlation:0.32548
  |
Correlation:0.32548
  |
Component:334; Jaccard:0.12072
 |
Component:087; Jaccard:0.13611
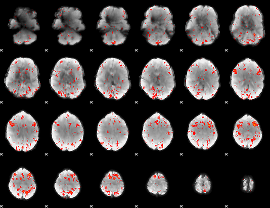 |
Component:334; Jaccard:0.15034
 |
Component:334; Jaccard:0.15725
 |
Component:334; Jaccard:0.15956
 |
Component:186; Jaccard:0.15593
 |
Component:186; Jaccard:0.1498
 |
Correlation:0.23761
  |
Correlation:0.073381
  |
Correlation:0.23761
  |
Correlation:0.23761
  |
Correlation:0.23761
  |
Correlation:0.17046
  |
Correlation:0.17046
  |
Cope: 02:"REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
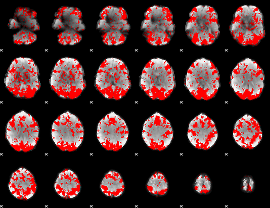 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
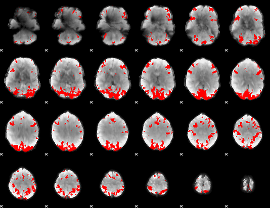 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
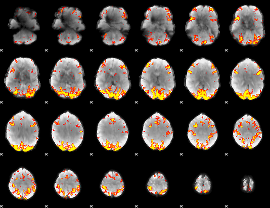 |
GLM Z-score result thresholded by 3.5
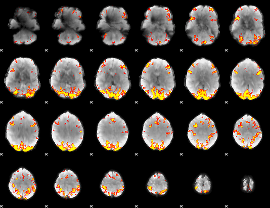 |
GLM Z-score result thresholded by 4
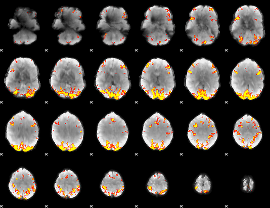 |
Component:070; Jaccard:0.17046
 |
Component:343; Jaccard:0.20605
 |
Component:343; Jaccard:0.25831
 |
Component:343; Jaccard:0.31107
 |
Component:343; Jaccard:0.36317
 |
Component:343; Jaccard:0.41034
 |
Component:343; Jaccard:0.44523
 |
Correlation:0.099668
  |
Correlation:0.33779
  |
Correlation:0.33779
  |
Correlation:0.33779
  |
Correlation:0.33779
  |
Correlation:0.33779
  |
Correlation:0.33779
  |
Component:343; Jaccard:0.16115
 |
Component:070; Jaccard:0.20132
 |
Component:070; Jaccard:0.22811
 |
Component:153; Jaccard:0.25412
 |
Component:153; Jaccard:0.27854
 |
Component:153; Jaccard:0.29531
 |
Component:153; Jaccard:0.30647
 |
Correlation:0.33779
  |
Correlation:0.099668
  |
Correlation:0.099668
  |
Correlation:0.13584
  |
Correlation:0.13584
  |
Correlation:0.13584
  |
Correlation:0.13584
  |
Component:375; Jaccard:0.15525
 |
Component:375; Jaccard:0.18927
 |
Component:375; Jaccard:0.22338
 |
Component:375; Jaccard:0.25244
 |
Component:375; Jaccard:0.27062
 |
Component:375; Jaccard:0.27893
 |
Component:375; Jaccard:0.2764
 |
Correlation:0.1759
  |
Correlation:0.1759
  |
Correlation:0.1759
  |
Correlation:0.1759
  |
Correlation:0.1759
  |
Correlation:0.1759
  |
Correlation:0.1759
  |
Component:250; Jaccard:0.15169
 |
Component:153; Jaccard:0.18561
 |
Component:153; Jaccard:0.22209
 |
Component:070; Jaccard:0.24558
 |
Component:070; Jaccard:0.25436
 |
Component:178; Jaccard:0.25927
 |
Component:178; Jaccard:0.26972
 |
Correlation:0.057298
  |
Correlation:0.13584
  |
Correlation:0.13584
  |
Correlation:0.099668
  |
Correlation:0.099668
  |
Correlation:0.3678
  |
Correlation:0.3678
  |
Component:153; Jaccard:0.15046
 |
Component:371; Jaccard:0.17562
 |
Component:371; Jaccard:0.2042
 |
Component:371; Jaccard:0.22795
 |
Component:371; Jaccard:0.24586
 |
Component:371; Jaccard:0.25682
 |
Component:371; Jaccard:0.25942
 |
Correlation:0.13584
  |
Correlation:0.30373
  |
Correlation:0.30373
  |
Correlation:0.30373
  |
Correlation:0.30373
  |
Correlation:0.30373
  |
Correlation:0.30373
  |
Component:371; Jaccard:0.14486
 |
Component:250; Jaccard:0.1711
 |
Component:225; Jaccard:0.19827
 |
Component:225; Jaccard:0.2227
 |
Component:178; Jaccard:0.2421
 |
Component:070; Jaccard:0.25442
 |
Component:225; Jaccard:0.25609
 |
Correlation:0.30373
  |
Correlation:0.057298
  |
Correlation:0.33861
  |
Correlation:0.33861
  |
Correlation:0.3678
  |
Correlation:0.099668
  |
Correlation:0.33861
  |
Component:225; Jaccard:0.13922
 |
Component:225; Jaccard:0.16906
 |
Component:178; Jaccard:0.19417
 |
Component:178; Jaccard:0.22055
 |
Component:225; Jaccard:0.24102
 |
Component:225; Jaccard:0.25262
 |
Component:070; Jaccard:0.24829
 |
Correlation:0.33861
  |
Correlation:0.33861
  |
Correlation:0.3678
  |
Correlation:0.3678
  |
Correlation:0.33861
  |
Correlation:0.33861
  |
Correlation:0.099668
  |
Component:178; Jaccard:0.13337
 |
Component:178; Jaccard:0.16416
 |
Component:250; Jaccard:0.18651
 |
Component:250; Jaccard:0.19276
 |
Component:250; Jaccard:0.19638
 |
Component:250; Jaccard:0.19282
 |
Component:250; Jaccard:0.18452
 |
Correlation:0.3678
  |
Correlation:0.3678
  |
Correlation:0.057298
  |
Correlation:0.057298
  |
Correlation:0.057298
  |
Correlation:0.057298
  |
Correlation:0.057298
  |
Component:087; Jaccard:0.12695
 |
Component:087; Jaccard:0.14287
 |
Component:087; Jaccard:0.15539
 |
Component:087; Jaccard:0.16341
 |
Component:087; Jaccard:0.16461
 |
Component:087; Jaccard:0.15903
 |
Component:087; Jaccard:0.14977
 |
Correlation:0.22651
  |
Correlation:0.22651
  |
Correlation:0.22651
  |
Correlation:0.22651
  |
Correlation:0.22651
  |
Correlation:0.22651
  |
Correlation:0.22651
  |
Component:275; Jaccard:0.12193
 |
Component:275; Jaccard:0.13595
 |
Component:275; Jaccard:0.14549
 |
Component:139; Jaccard:0.14988
 |
Component:139; Jaccard:0.14938
 |
Component:139; Jaccard:0.14386
 |
Component:186; Jaccard:0.13842
 |
Correlation:0.1699
  |
Correlation:0.1699
  |
Correlation:0.1699
  |
Correlation:0.092096
  |
Correlation:0.092096
  |
Correlation:0.092096
  |
Correlation:0.031928
  |
Cope: 03:"MATCH-REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
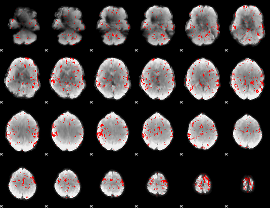 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
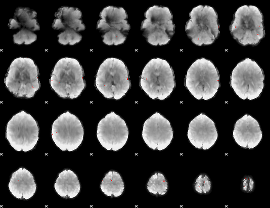 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
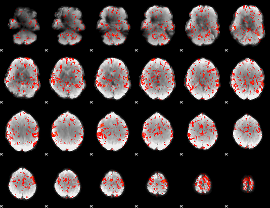 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
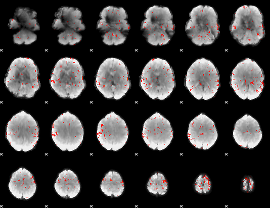 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
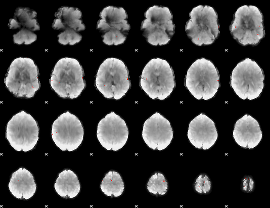 |
Component:146; Jaccard:0.12866
 |
Component:146; Jaccard:0.13609
 |
Component:146; Jaccard:0.12263
 |
Component:146; Jaccard:0.084464
 |
Component:146; Jaccard:0.044607
 |
Component:146; Jaccard:0.017351
 |
Component:146; Jaccard:0.005748
 |
Correlation:0.29358
  |
Correlation:0.29358
  |
Correlation:0.29358
  |
Correlation:0.29358
  |
Correlation:0.29358
  |
Correlation:0.29358
  |
Correlation:0.29358
  |
Component:204; Jaccard:0.11303
 |
Component:204; Jaccard:0.11575
 |
Component:204; Jaccard:0.09754
 |
Component:074; Jaccard:0.063594
 |
Component:074; Jaccard:0.035146
 |
Component:074; Jaccard:0.013749
 |
Component:149; Jaccard:0.004729
 |
Correlation:0.13574
  |
Correlation:0.13574
  |
Correlation:0.13574
  |
Correlation:0.23879
  |
Correlation:0.23879
  |
Correlation:0.23879
  |
Correlation:0.13827
  |
Component:184; Jaccard:0.10339
 |
Component:074; Jaccard:0.10184
 |
Component:074; Jaccard:0.093588
 |
Component:204; Jaccard:0.062006
 |
Component:204; Jaccard:0.031255
 |
Component:227; Jaccard:0.01169
 |
Component:074; Jaccard:0.004471
 |
Correlation:0.18116
  |
Correlation:0.23879
  |
Correlation:0.23879
  |
Correlation:0.13574
  |
Correlation:0.13574
  |
Correlation:0.28096
  |
Correlation:0.23879
  |
Component:074; Jaccard:0.095716
 |
Component:184; Jaccard:0.09759
 |
Component:184; Jaccard:0.077477
 |
Component:282; Jaccard:0.050265
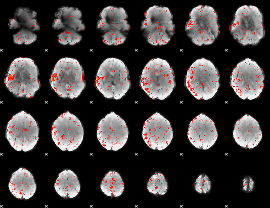 |
Component:282; Jaccard:0.026696
 |
Component:204; Jaccard:0.011226
 |
Component:244; Jaccard:0.003966
 |
Correlation:0.23879
  |
Correlation:0.18116
  |
Correlation:0.18116
  |
Correlation:0.24861
  |
Correlation:0.24861
  |
Correlation:0.13574
  |
Correlation:0.15613
  |
Component:244; Jaccard:0.086525
 |
Component:244; Jaccard:0.085278
 |
Component:249; Jaccard:0.073223
 |
Component:249; Jaccard:0.049968
 |
Component:244; Jaccard:0.024482
 |
Component:282; Jaccard:0.011206
 |
Component:365; Jaccard:0.003918
 |
Correlation:0.15613
  |
Correlation:0.15613
  |
Correlation:0.2108
  |
Correlation:0.2108
  |
Correlation:0.15613
  |
Correlation:0.24861
  |
Correlation:0.12122
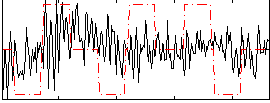 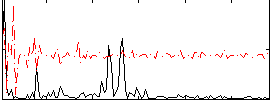 |
Component:249; Jaccard:0.082781
 |
Component:249; Jaccard:0.083757
 |
Component:282; Jaccard:0.072202
 |
Component:244; Jaccard:0.046944
 |
Component:227; Jaccard:0.024335
 |
Component:244; Jaccard:0.010581
 |
Component:268; Jaccard:0.003394
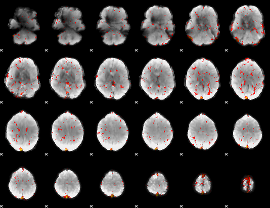 |
Correlation:0.2108
  |
Correlation:0.2108
  |
Correlation:0.24861
  |
Correlation:0.15613
  |
Correlation:0.28096
  |
Correlation:0.15613
  |
Correlation:0.10591
  |
Component:282; Jaccard:0.079828
 |
Component:282; Jaccard:0.079087
 |
Component:244; Jaccard:0.068967
 |
Component:184; Jaccard:0.046756
 |
Component:268; Jaccard:0.02226
 |
Component:149; Jaccard:0.010257
 |
Component:028; Jaccard:0.003051
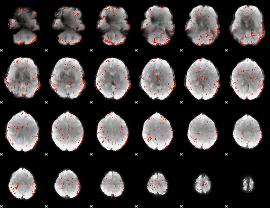 |
Correlation:0.24861
  |
Correlation:0.24861
  |
Correlation:0.15613
  |
Correlation:0.18116
  |
Correlation:0.10591
  |
Correlation:0.13827
  |
Correlation:0.21134
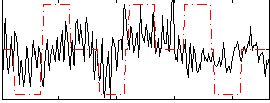  |
Component:259; Jaccard:0.079017
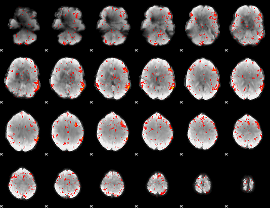 |
Component:259; Jaccard:0.073332
 |
Component:004; Jaccard:0.060182
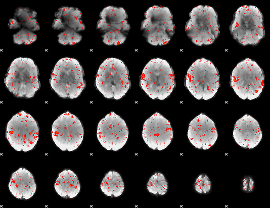 |
Component:227; Jaccard:0.042843
 |
Component:249; Jaccard:0.022079
 |
Component:268; Jaccard:0.009915
 |
Component:204; Jaccard:0.003037
 |
Correlation:0.15982
  |
Correlation:0.15982
  |
Correlation:0.13286
  |
Correlation:0.28096
  |
Correlation:0.2108
  |
Correlation:0.10591
  |
Correlation:0.13574
  |
Component:004; Jaccard:0.073793
 |
Component:004; Jaccard:0.07268
 |
Component:259; Jaccard:0.05849
 |
Component:268; Jaccard:0.038939
 |
Component:184; Jaccard:0.020683
 |
Component:269; Jaccard:0.009623
 |
Component:282; Jaccard:0.002984
 |
Correlation:0.13286
  |
Correlation:0.13286
  |
Correlation:0.15982
  |
Correlation:0.10591
  |
Correlation:0.18116
  |
Correlation:0.12507
  |
Correlation:0.24861
  |
Component:034; Jaccard:0.071594
 |
Component:183; Jaccard:0.066109
 |
Component:227; Jaccard:0.055215
 |
Component:004; Jaccard:0.038275
 |
Component:149; Jaccard:0.020336
 |
Component:365; Jaccard:0.009266
 |
Component:227; Jaccard:0.002902
 |
Correlation:0.076505
  |
Correlation:0.014203
  |
Correlation:0.28096
  |
Correlation:0.13286
  |
Correlation:0.13827
  |
Correlation:0.12122
  |
Correlation:0.28096
  |
Cope: 04:"REL-MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
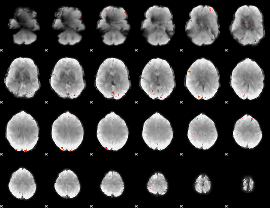 |
Component:343; Jaccard:0.12629
 |
Component:343; Jaccard:0.15287
 |
Component:343; Jaccard:0.16255
 |
Component:343; Jaccard:0.14508
 |
Component:343; Jaccard:0.10612
 |
Component:343; Jaccard:0.062853
 |
Component:343; Jaccard:0.034
 |
Correlation:0.10348
 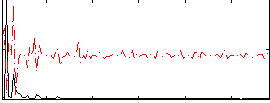 |
Correlation:0.10348
  |
Correlation:0.10348
  |
Correlation:0.10348
  |
Correlation:0.10348
  |
Correlation:0.10348
  |
Correlation:0.10348
  |
Component:367; Jaccard:0.12399
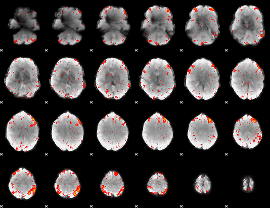 |
Component:367; Jaccard:0.13627
 |
Component:178; Jaccard:0.145
 |
Component:178; Jaccard:0.13079
 |
Component:178; Jaccard:0.092661
 |
Component:178; Jaccard:0.054834
 |
Component:178; Jaccard:0.0303
 |
Correlation:0.012131
 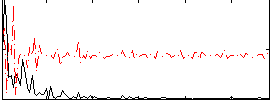 |
Correlation:0.012131
  |
Correlation:0.15579
  |
Correlation:0.15579
  |
Correlation:0.15579
  |
Correlation:0.15579
  |
Correlation:0.15579
  |
Component:178; Jaccard:0.10879
 |
Component:178; Jaccard:0.1319
 |
Component:367; Jaccard:0.13359
 |
Component:367; Jaccard:0.10648
 |
Component:367; Jaccard:0.067746
 |
Component:225; Jaccard:0.036459
 |
Component:225; Jaccard:0.019985
 |
Correlation:0.15579
  |
Correlation:0.15579
  |
Correlation:0.012131
  |
Correlation:0.012131
  |
Correlation:0.012131
  |
Correlation:0.10914
  |
Correlation:0.10914
  |
Component:225; Jaccard:0.10034
 |
Component:225; Jaccard:0.11212
 |
Component:225; Jaccard:0.10981
 |
Component:225; Jaccard:0.091746
 |
Component:225; Jaccard:0.063896
 |
Component:367; Jaccard:0.035272
 |
Component:367; Jaccard:0.01822
 |
Correlation:0.10914
  |
Correlation:0.10914
  |
Correlation:0.10914
  |
Correlation:0.10914
  |
Correlation:0.10914
  |
Correlation:0.012131
  |
Correlation:0.012131
  |
Component:099; Jaccard:0.0961
 |
Component:041; Jaccard:0.10162
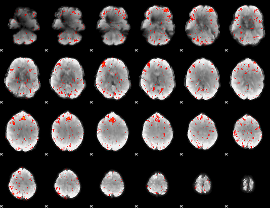 |
Component:348; Jaccard:0.10163
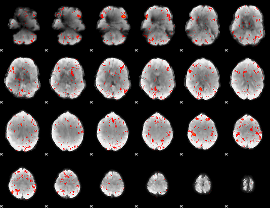 |
Component:348; Jaccard:0.08575
 |
Component:348; Jaccard:0.055428
 |
Component:070; Jaccard:0.031994
 |
Component:070; Jaccard:0.017704
 |
Correlation:0.12541
  |
Correlation:0.071295
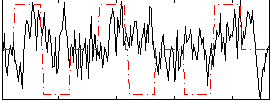  |
Correlation:0.14206
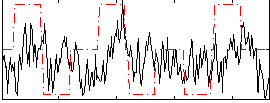  |
Correlation:0.14206
  |
Correlation:0.14206
  |
Correlation:-0.012919
  |
Correlation:-0.012919
  |
Component:052; Jaccard:0.09414
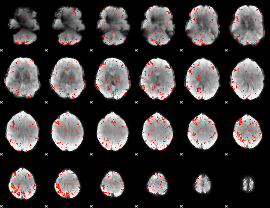 |
Component:052; Jaccard:0.10069
 |
Component:041; Jaccard:0.098615
 |
Component:070; Jaccard:0.077427
 |
Component:070; Jaccard:0.054822
 |
Component:348; Jaccard:0.028972
 |
Component:153; Jaccard:0.015528
 |
Correlation:0.067515
  |
Correlation:0.067515
  |
Correlation:0.071295
  |
Correlation:-0.012919
  |
Correlation:-0.012919
  |
Correlation:0.14206
  |
Correlation:-0.032687
 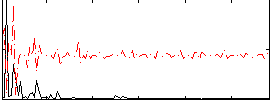 |
Component:041; Jaccard:0.0932
 |
Component:348; Jaccard:0.10062
 |
Component:070; Jaccard:0.094087
 |
Component:041; Jaccard:0.076529
 |
Component:041; Jaccard:0.050671
 |
Component:153; Jaccard:0.026103
 |
Component:348; Jaccard:0.014149
 |
Correlation:0.071295
  |
Correlation:0.14206
  |
Correlation:-0.012919
  |
Correlation:0.071295
  |
Correlation:0.071295
  |
Correlation:-0.032687
  |
Correlation:0.14206
  |
Component:070; Jaccard:0.092142
 |
Component:070; Jaccard:0.097874
 |
Component:052; Jaccard:0.093642
 |
Component:052; Jaccard:0.071537
 |
Component:153; Jaccard:0.044713
 |
Component:041; Jaccard:0.025941
 |
Component:041; Jaccard:0.011505
 |
Correlation:-0.012919
  |
Correlation:-0.012919
  |
Correlation:0.067515
  |
Correlation:0.067515
  |
Correlation:-0.032687
  |
Correlation:0.071295
  |
Correlation:0.071295
  |
Component:348; Jaccard:0.091423
 |
Component:099; Jaccard:0.096017
 |
Component:099; Jaccard:0.083999
 |
Component:153; Jaccard:0.064427
 |
Component:052; Jaccard:0.044486
 |
Component:052; Jaccard:0.022152
 |
Component:052; Jaccard:0.009488
 |
Correlation:0.14206
  |
Correlation:0.12541
  |
Correlation:0.12541
  |
Correlation:-0.032687
  |
Correlation:0.067515
  |
Correlation:0.067515
  |
Correlation:0.067515
  |
Component:293; Jaccard:0.088956
 |
Component:293; Jaccard:0.087594
 |
Component:153; Jaccard:0.079098
 |
Component:099; Jaccard:0.057846
 |
Component:371; Jaccard:0.035234
 |
Component:371; Jaccard:0.017602
 |
Component:250; Jaccard:0.007946
 |
Correlation:0.10679
 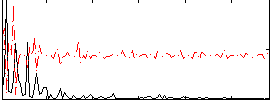 |
Correlation:0.10679
  |
Correlation:-0.032687
  |
Correlation:0.12541
  |
Correlation:0.12977
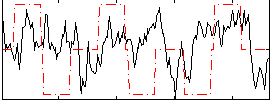  |
Correlation:0.12977
  |
Correlation:-0.0063068
 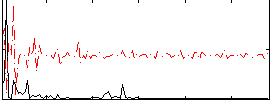 |
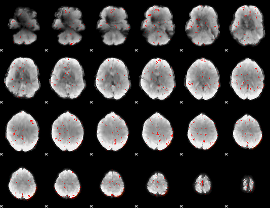
Cope: 05:"neg_MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
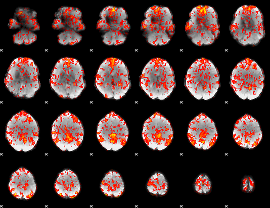 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
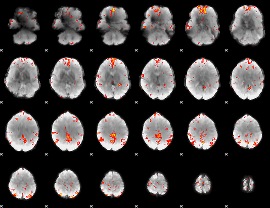 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:389; Jaccard:0.15167
 |
Component:389; Jaccard:0.19827
 |
Component:389; Jaccard:0.25358
 |
Component:389; Jaccard:0.30001
 |
Component:389; Jaccard:0.30451
 |
Component:389; Jaccard:0.26689
 |
Component:389; Jaccard:0.20826
 |
Correlation:0.030943
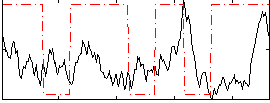  |
Correlation:0.030943
  |
Correlation:0.030943
  |
Correlation:0.030943
  |
Correlation:0.030943
  |
Correlation:0.030943
  |
Correlation:0.030943
  |
Component:249; Jaccard:0.11611
 |
Component:249; Jaccard:0.13536
 |
Component:249; Jaccard:0.15112
 |
Component:249; Jaccard:0.15689
 |
Component:249; Jaccard:0.14562
 |
Component:249; Jaccard:0.11728
 |
Component:249; Jaccard:0.08453
 |
Correlation:0.083504
  |
Correlation:0.083504
  |
Correlation:0.083504
  |
Correlation:0.083504
  |
Correlation:0.083504
  |
Correlation:0.083504
  |
Correlation:0.083504
  |
Component:189; Jaccard:0.11247
 |
Component:125; Jaccard:0.12644
 |
Component:125; Jaccard:0.1378
 |
Component:125; Jaccard:0.13654
 |
Component:065; Jaccard:0.12773
 |
Component:065; Jaccard:0.10951
 |
Component:065; Jaccard:0.084027
 |
Correlation:0.094554
  |
Correlation:0.090993
  |
Correlation:0.090993
  |
Correlation:0.090993
  |
Correlation:0.14251
  |
Correlation:0.14251
  |
Correlation:0.14251
  |
Component:125; Jaccard:0.10971
 |
Component:189; Jaccard:0.12573
 |
Component:189; Jaccard:0.1325
 |
Component:065; Jaccard:0.13112
 |
Component:125; Jaccard:0.12085
 |
Component:125; Jaccard:0.097013
 |
Component:125; Jaccard:0.071358
 |
Correlation:0.090993
  |
Correlation:0.094554
  |
Correlation:0.094554
  |
Correlation:0.14251
  |
Correlation:0.090993
  |
Correlation:0.090993
  |
Correlation:0.090993
  |
Component:016; Jaccard:0.10501
 |
Component:156; Jaccard:0.11558
 |
Component:156; Jaccard:0.1252
 |
Component:189; Jaccard:0.12903
 |
Component:189; Jaccard:0.11509
 |
Component:156; Jaccard:0.08731
 |
Component:190; Jaccard:0.067212
 |
Correlation:0.1838
  |
Correlation:0.099136
  |
Correlation:0.099136
  |
Correlation:0.094554
  |
Correlation:0.094554
  |
Correlation:0.099136
  |
Correlation:-0.12288
  |
Component:074; Jaccard:0.10358
 |
Component:016; Jaccard:0.11555
 |
Component:065; Jaccard:0.1233
 |
Component:156; Jaccard:0.12477
 |
Component:156; Jaccard:0.11202
 |
Component:189; Jaccard:0.086823
 |
Component:189; Jaccard:0.062369
 |
Correlation:-0.099905
  |
Correlation:0.1838
  |
Correlation:0.14251
  |
Correlation:0.099136
  |
Correlation:0.099136
  |
Correlation:0.094554
  |
Correlation:0.094554
  |
Component:156; Jaccard:0.10298
 |
Component:074; Jaccard:0.11219
 |
Component:016; Jaccard:0.12014
 |
Component:016; Jaccard:0.1148
 |
Component:016; Jaccard:0.10317
 |
Component:190; Jaccard:0.083208
 |
Component:156; Jaccard:0.06102
 |
Correlation:0.099136
  |
Correlation:-0.099905
  |
Correlation:0.1838
  |
Correlation:0.1838
  |
Correlation:0.1838
  |
Correlation:-0.12288
  |
Correlation:0.099136
  |
Component:184; Jaccard:0.098902
 |
Component:065; Jaccard:0.10655
 |
Component:074; Jaccard:0.11421
 |
Component:074; Jaccard:0.10875
 |
Component:190; Jaccard:0.096979
 |
Component:016; Jaccard:0.079633
 |
Component:016; Jaccard:0.055346
 |
Correlation:0.041071
  |
Correlation:0.14251
  |
Correlation:-0.099905
  |
Correlation:-0.099905
  |
Correlation:-0.12288
  |
Correlation:0.1838
  |
Correlation:0.1838
  |
Component:195; Jaccard:0.089834
 |
Component:195; Jaccard:0.10065
 |
Component:195; Jaccard:0.10632
 |
Component:195; Jaccard:0.10582
 |
Component:195; Jaccard:0.093448
 |
Component:074; Jaccard:0.07211
 |
Component:299; Jaccard:0.052854
 |
Correlation:-0.031017
  |
Correlation:-0.031017
  |
Correlation:-0.031017
  |
Correlation:-0.031017
  |
Correlation:-0.031017
  |
Correlation:-0.099905
  |
Correlation:0.19275
  |
Component:065; Jaccard:0.089086
 |
Component:184; Jaccard:0.099082
 |
Component:315; Jaccard:0.10135
 |
Component:190; Jaccard:0.10105
 |
Component:074; Jaccard:0.092232
 |
Component:315; Jaccard:0.070502
 |
Component:074; Jaccard:0.051843
 |
Correlation:0.14251
  |
Correlation:0.041071
  |
Correlation:-0.088423
  |
Correlation:-0.12288
  |
Correlation:-0.099905
  |
Correlation:-0.088423
  |
Correlation:-0.099905
  |
Cope: 06:"neg_REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
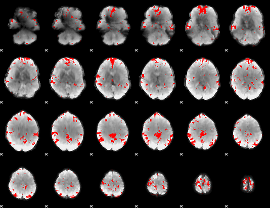 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
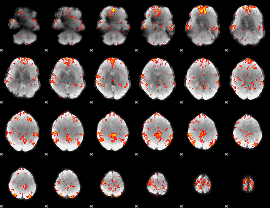 |
GLM Z-score result thresholded by 3
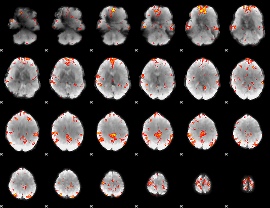 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:389; Jaccard:0.15165
 |
Component:389; Jaccard:0.19906
 |
Component:389; Jaccard:0.2563
 |
Component:389; Jaccard:0.3029
 |
Component:389; Jaccard:0.32652
 |
Component:389; Jaccard:0.31105
 |
Component:389; Jaccard:0.26518
 |
Correlation:0.37714
  |
Correlation:0.37714
  |
Correlation:0.37714
  |
Correlation:0.37714
  |
Correlation:0.37714
  |
Correlation:0.37714
  |
Correlation:0.37714
  |
Component:249; Jaccard:0.13188
 |
Component:249; Jaccard:0.16029
 |
Component:249; Jaccard:0.18735
 |
Component:249; Jaccard:0.2023
 |
Component:249; Jaccard:0.20033
 |
Component:249; Jaccard:0.17976
 |
Component:249; Jaccard:0.14902
 |
Correlation:0.43917
  |
Correlation:0.43917
  |
Correlation:0.43917
  |
Correlation:0.43917
  |
Correlation:0.43917
  |
Correlation:0.43917
  |
Correlation:0.43917
  |
Component:184; Jaccard:0.1277
 |
Component:074; Jaccard:0.14583
 |
Component:074; Jaccard:0.16034
 |
Component:074; Jaccard:0.16545
 |
Component:074; Jaccard:0.15623
 |
Component:074; Jaccard:0.13631
 |
Component:074; Jaccard:0.10725
 |
Correlation:0.34672
  |
Correlation:0.30297
  |
Correlation:0.30297
  |
Correlation:0.30297
  |
Correlation:0.30297
  |
Correlation:0.30297
  |
Correlation:0.30297
  |
Component:074; Jaccard:0.12614
 |
Component:189; Jaccard:0.14276
 |
Component:189; Jaccard:0.15442
 |
Component:189; Jaccard:0.15576
 |
Component:189; Jaccard:0.14434
 |
Component:189; Jaccard:0.12141
 |
Component:189; Jaccard:0.098074
 |
Correlation:0.30297
  |
Correlation:0.30739
  |
Correlation:0.30739
  |
Correlation:0.30739
  |
Correlation:0.30739
  |
Correlation:0.30739
  |
Correlation:0.30739
  |
Component:189; Jaccard:0.12378
 |
Component:184; Jaccard:0.13825
 |
Component:184; Jaccard:0.14251
 |
Component:184; Jaccard:0.13433
 |
Component:065; Jaccard:0.12639
 |
Component:065; Jaccard:0.11697
 |
Component:065; Jaccard:0.096291
 |
Correlation:0.30739
  |
Correlation:0.34672
  |
Correlation:0.34672
  |
Correlation:0.34672
  |
Correlation:0.25752
  |
Correlation:0.25752
  |
Correlation:0.25752
  |
Component:125; Jaccard:0.10116
 |
Component:125; Jaccard:0.11377
 |
Component:156; Jaccard:0.12317
 |
Component:065; Jaccard:0.12745
 |
Component:184; Jaccard:0.1152
 |
Component:190; Jaccard:0.10042
 |
Component:190; Jaccard:0.085349
 |
Correlation:0.22963
  |
Correlation:0.22963
  |
Correlation:0.3633
  |
Correlation:0.25752
  |
Correlation:0.34672
  |
Correlation:0.38493
  |
Correlation:0.38493
  |
Component:156; Jaccard:0.098584
 |
Component:156; Jaccard:0.11252
 |
Component:125; Jaccard:0.12292
 |
Component:156; Jaccard:0.12441
 |
Component:125; Jaccard:0.11466
 |
Component:195; Jaccard:0.10003
 |
Component:195; Jaccard:0.080722
 |
Correlation:0.3633
  |
Correlation:0.3633
  |
Correlation:0.22963
  |
Correlation:0.3633
  |
Correlation:0.22963
  |
Correlation:0.33919
  |
Correlation:0.33919
  |
Component:204; Jaccard:0.098403
 |
Component:195; Jaccard:0.10848
 |
Component:065; Jaccard:0.12163
 |
Component:125; Jaccard:0.12432
 |
Component:195; Jaccard:0.11453
 |
Component:125; Jaccard:0.096644
 |
Component:125; Jaccard:0.075091
 |
Correlation:0.29385
  |
Correlation:0.33919
  |
Correlation:0.25752
  |
Correlation:0.22963
  |
Correlation:0.33919
  |
Correlation:0.22963
  |
Correlation:0.22963
  |
Component:195; Jaccard:0.096572
 |
Component:065; Jaccard:0.10695
 |
Component:195; Jaccard:0.11741
 |
Component:195; Jaccard:0.1196
 |
Component:156; Jaccard:0.11389
 |
Component:156; Jaccard:0.095522
 |
Component:156; Jaccard:0.073033
 |
Correlation:0.33919
  |
Correlation:0.25752
  |
Correlation:0.33919
  |
Correlation:0.33919
  |
Correlation:0.3633
  |
Correlation:0.3633
  |
Correlation:0.3633
  |
Component:315; Jaccard:0.094808
 |
Component:315; Jaccard:0.10557
 |
Component:315; Jaccard:0.11318
 |
Component:190; Jaccard:0.11124
 |
Component:190; Jaccard:0.11084
 |
Component:184; Jaccard:0.092032
 |
Component:315; Jaccard:0.06646
 |
Correlation:0.23967
  |
Correlation:0.23967
  |
Correlation:0.23967
  |
Correlation:0.38493
  |
Correlation:0.38493
  |
Correlation:0.34672
  |
Correlation:0.23967
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































