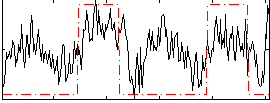
Task name: GAMBLING, TR=0.72, totally 253 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"PUNISH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
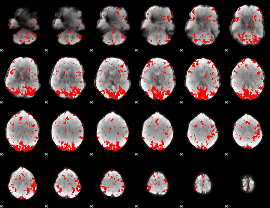 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
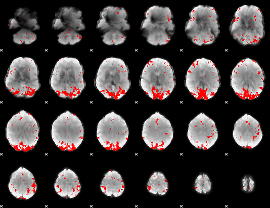 |
GLM binary result thresholded by 3.5
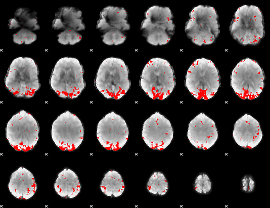 |
GLM binary result thresholded by 4
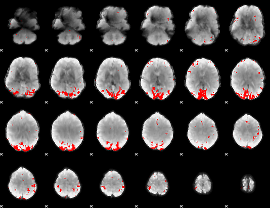 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
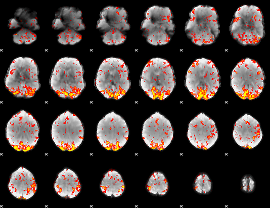 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
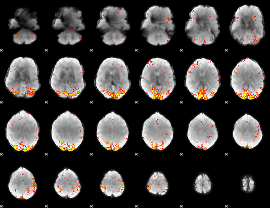 |
GLM Z-score result thresholded by 4
 |
Component:157; Jaccard:0.14902
 |
Component:157; Jaccard:0.1947
 |
Component:157; Jaccard:0.24994
 |
Component:157; Jaccard:0.30385
 |
Component:157; Jaccard:0.34481
 |
Component:157; Jaccard:0.35696
 |
Component:157; Jaccard:0.34694
 |
Correlation:0.2513
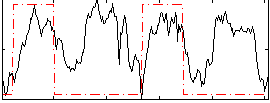 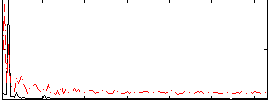 |
Correlation:0.2513
  |
Correlation:0.2513
  |
Correlation:0.2513
  |
Correlation:0.2513
  |
Correlation:0.2513
  |
Correlation:0.2513
  |
Component:315; Jaccard:0.13478
 |
Component:315; Jaccard:0.16474
 |
Component:315; Jaccard:0.19306
 |
Component:315; Jaccard:0.2137
 |
Component:315; Jaccard:0.22135
 |
Component:315; Jaccard:0.21362
 |
Component:153; Jaccard:0.20621
 |
Correlation:0.42653
  |
Correlation:0.42653
  |
Correlation:0.42653
  |
Correlation:0.42653
  |
Correlation:0.42653
  |
Correlation:0.42653
  |
Correlation:0.38606
  |
Component:170; Jaccard:0.098649
 |
Component:153; Jaccard:0.12071
 |
Component:153; Jaccard:0.14697
 |
Component:153; Jaccard:0.17311
 |
Component:153; Jaccard:0.19173
 |
Component:153; Jaccard:0.20367
 |
Component:315; Jaccard:0.19669
 |
Correlation:0.39238
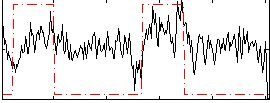 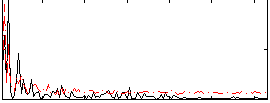 |
Correlation:0.38606
  |
Correlation:0.38606
  |
Correlation:0.38606
  |
Correlation:0.38606
  |
Correlation:0.38606
  |
Correlation:0.42653
  |
Component:153; Jaccard:0.095972
 |
Component:397; Jaccard:0.1176
 |
Component:397; Jaccard:0.14101
 |
Component:397; Jaccard:0.16113
 |
Component:397; Jaccard:0.1688
 |
Component:397; Jaccard:0.16278
 |
Component:397; Jaccard:0.15045
 |
Correlation:0.38606
  |
Correlation:0.32215
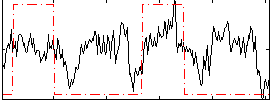  |
Correlation:0.32215
  |
Correlation:0.32215
  |
Correlation:0.32215
  |
Correlation:0.32215
  |
Correlation:0.32215
  |
Component:136; Jaccard:0.095752
 |
Component:136; Jaccard:0.11416
 |
Component:136; Jaccard:0.13299
 |
Component:136; Jaccard:0.14729
 |
Component:136; Jaccard:0.15212
 |
Component:136; Jaccard:0.14724
 |
Component:136; Jaccard:0.13605
 |
Correlation:0.22553
  |
Correlation:0.22553
  |
Correlation:0.22553
  |
Correlation:0.22553
  |
Correlation:0.22553
  |
Correlation:0.22553
  |
Correlation:0.22553
  |
Component:397; Jaccard:0.095369
 |
Component:283; Jaccard:0.11244
 |
Component:283; Jaccard:0.12635
 |
Component:283; Jaccard:0.13722
 |
Component:283; Jaccard:0.13892
 |
Component:283; Jaccard:0.13414
 |
Component:318; Jaccard:0.12462
 |
Correlation:0.32215
  |
Correlation:0.53155
 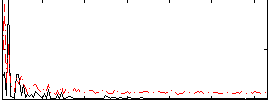 |
Correlation:0.53155
  |
Correlation:0.53155
  |
Correlation:0.53155
  |
Correlation:0.53155
  |
Correlation:0.41908
  |
Component:283; Jaccard:0.095212
 |
Component:170; Jaccard:0.10994
 |
Component:318; Jaccard:0.12351
 |
Component:318; Jaccard:0.13085
 |
Component:318; Jaccard:0.13373
 |
Component:318; Jaccard:0.12917
 |
Component:283; Jaccard:0.12439
 |
Correlation:0.53155
  |
Correlation:0.39238
  |
Correlation:0.41908
  |
Correlation:0.41908
  |
Correlation:0.41908
  |
Correlation:0.41908
  |
Correlation:0.53155
  |
Component:318; Jaccard:0.093885
 |
Component:318; Jaccard:0.10938
 |
Component:170; Jaccard:0.11716
 |
Component:170; Jaccard:0.11803
 |
Component:265; Jaccard:0.11968
 |
Component:265; Jaccard:0.11624
 |
Component:265; Jaccard:0.10719
 |
Correlation:0.41908
  |
Correlation:0.41908
  |
Correlation:0.39238
  |
Correlation:0.39238
  |
Correlation:0.19835
  |
Correlation:0.19835
  |
Correlation:0.19835
  |
Component:393; Jaccard:0.091629
 |
Component:393; Jaccard:0.10273
 |
Component:393; Jaccard:0.11239
 |
Component:265; Jaccard:0.11642
 |
Component:393; Jaccard:0.11399
 |
Component:249; Jaccard:0.10629
 |
Component:170; Jaccard:0.096335
 |
Correlation:0.097406
  |
Correlation:0.097406
  |
Correlation:0.097406
  |
Correlation:0.19835
  |
Correlation:0.097406
  |
Correlation:0.30323
  |
Correlation:0.39238
  |
Component:201; Jaccard:0.088044
 |
Component:201; Jaccard:0.097803
 |
Component:265; Jaccard:0.10789
 |
Component:393; Jaccard:0.116
 |
Component:170; Jaccard:0.11246
 |
Component:170; Jaccard:0.10498
 |
Component:314; Jaccard:0.095761
 |
Correlation:0.29865
  |
Correlation:0.29865
  |
Correlation:0.19835
  |
Correlation:0.097406
  |
Correlation:0.39238
  |
Correlation:0.39238
  |
Correlation:0.086671
  |
Cope: 02:"REWARD" BACK
|
GLM result group-wise
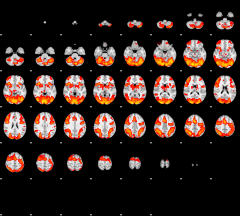 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
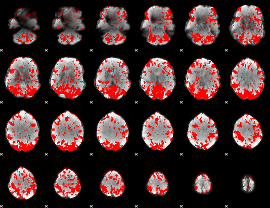 |
GLM binary result thresholded by 2
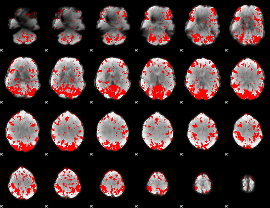 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
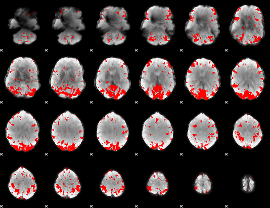 |
GLM binary result thresholded by 3.5
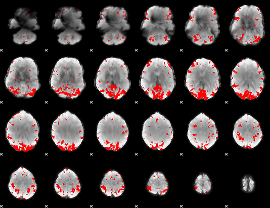 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
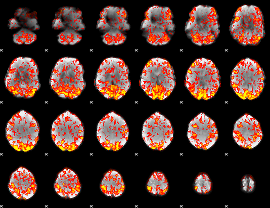 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
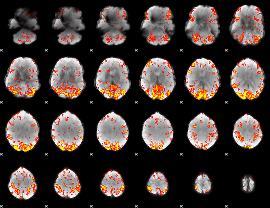 |
GLM Z-score result thresholded by 3
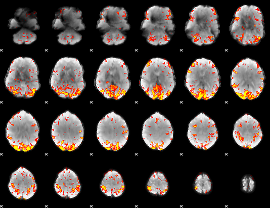 |
GLM Z-score result thresholded by 3.5
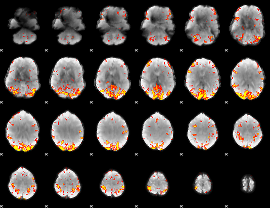 |
GLM Z-score result thresholded by 4
 |
Component:315; Jaccard:0.12155
 |
Component:157; Jaccard:0.14799
 |
Component:157; Jaccard:0.18305
 |
Component:157; Jaccard:0.22313
 |
Component:157; Jaccard:0.26419
 |
Component:157; Jaccard:0.29988
 |
Component:157; Jaccard:0.32267
 |
Correlation:0.082254
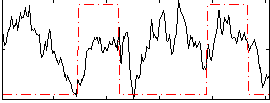  |
Correlation:0.41637
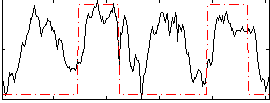 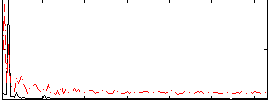 |
Correlation:0.41637
  |
Correlation:0.41637
  |
Correlation:0.41637
  |
Correlation:0.41637
  |
Correlation:0.41637
  |
Component:157; Jaccard:0.119
 |
Component:315; Jaccard:0.1464
 |
Component:315; Jaccard:0.17169
 |
Component:315; Jaccard:0.19539
 |
Component:315; Jaccard:0.21771
 |
Component:315; Jaccard:0.2272
 |
Component:315; Jaccard:0.22972
 |
Correlation:0.41637
  |
Correlation:0.082254
  |
Correlation:0.082254
  |
Correlation:0.082254
  |
Correlation:0.082254
  |
Correlation:0.082254
  |
Correlation:0.082254
  |
Component:393; Jaccard:0.10399
 |
Component:393; Jaccard:0.124
 |
Component:393; Jaccard:0.14365
 |
Component:393; Jaccard:0.16058
 |
Component:393; Jaccard:0.17133
 |
Component:397; Jaccard:0.18113
 |
Component:397; Jaccard:0.19023
 |
Correlation:0.063313
  |
Correlation:0.063313
  |
Correlation:0.063313
  |
Correlation:0.063313
  |
Correlation:0.063313
  |
Correlation:0.22673
  |
Correlation:0.22673
  |
Component:232; Jaccard:0.094813
 |
Component:232; Jaccard:0.10297
 |
Component:397; Jaccard:0.12234
 |
Component:397; Jaccard:0.14417
 |
Component:397; Jaccard:0.16531
 |
Component:393; Jaccard:0.17348
 |
Component:393; Jaccard:0.16425
 |
Correlation:0.028974
  |
Correlation:0.028974
  |
Correlation:0.22673
  |
Correlation:0.22673
  |
Correlation:0.22673
  |
Correlation:0.063313
  |
Correlation:0.063313
  |
Component:115; Jaccard:0.088081
 |
Component:136; Jaccard:0.10167
 |
Component:136; Jaccard:0.11905
 |
Component:136; Jaccard:0.13524
 |
Component:136; Jaccard:0.14866
 |
Component:136; Jaccard:0.15658
 |
Component:136; Jaccard:0.15662
 |
Correlation:-0.13358
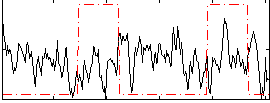 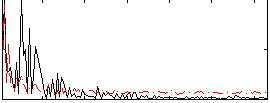 |
Correlation:0.19441
  |
Correlation:0.19441
  |
Correlation:0.19441
  |
Correlation:0.19441
  |
Correlation:0.19441
  |
Correlation:0.19441
  |
Component:211; Jaccard:0.086428
 |
Component:397; Jaccard:0.10134
 |
Component:211; Jaccard:0.11012
 |
Component:211; Jaccard:0.11567
 |
Component:153; Jaccard:0.12778
 |
Component:153; Jaccard:0.13995
 |
Component:153; Jaccard:0.1513
 |
Correlation:0.2046
  |
Correlation:0.22673
  |
Correlation:0.2046
  |
Correlation:0.2046
  |
Correlation:0.18375
  |
Correlation:0.18375
  |
Correlation:0.18375
  |
Component:136; Jaccard:0.086203
 |
Component:211; Jaccard:0.098549
 |
Component:232; Jaccard:0.10937
 |
Component:232; Jaccard:0.11332
 |
Component:211; Jaccard:0.11971
 |
Component:314; Jaccard:0.12545
 |
Component:314; Jaccard:0.12637
 |
Correlation:0.19441
  |
Correlation:0.2046
  |
Correlation:0.028974
  |
Correlation:0.028974
  |
Correlation:0.2046
  |
Correlation:0.43444
  |
Correlation:0.43444
  |
Component:249; Jaccard:0.083694
 |
Component:115; Jaccard:0.095819
 |
Component:115; Jaccard:0.10086
 |
Component:153; Jaccard:0.1132
 |
Component:314; Jaccard:0.11847
 |
Component:211; Jaccard:0.12027
 |
Component:283; Jaccard:0.11708
 |
Correlation:0.10265
  |
Correlation:-0.13358
  |
Correlation:-0.13358
  |
Correlation:0.18375
  |
Correlation:0.43444
  |
Correlation:0.2046
  |
Correlation:0.059457
  |
Component:397; Jaccard:0.083348
 |
Component:249; Jaccard:0.091999
 |
Component:314; Jaccard:0.10084
 |
Component:314; Jaccard:0.1109
 |
Component:283; Jaccard:0.11198
 |
Component:283; Jaccard:0.11718
 |
Component:211; Jaccard:0.11619
 |
Correlation:0.22673
  |
Correlation:0.10265
  |
Correlation:0.43444
  |
Correlation:0.43444
  |
Correlation:0.059457
  |
Correlation:0.059457
  |
Correlation:0.2046
  |
Component:314; Jaccard:0.079343
 |
Component:314; Jaccard:0.090527
 |
Component:249; Jaccard:0.10007
 |
Component:249; Jaccard:0.10555
 |
Component:232; Jaccard:0.11141
 |
Component:249; Jaccard:0.11185
 |
Component:318; Jaccard:0.11155
 |
Correlation:0.43444
  |
Correlation:0.43444
  |
Correlation:0.10265
  |
Correlation:0.10265
  |
Correlation:0.028974
  |
Correlation:0.10265
  |
Correlation:-0.060422
  |
Cope: 03:"PUNISH-REWARD" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
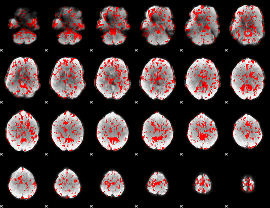 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
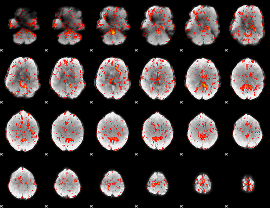 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
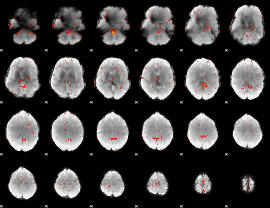 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:306; Jaccard:0.11622
 |
Component:306; Jaccard:0.1398
 |
Component:306; Jaccard:0.15849
 |
Component:306; Jaccard:0.16462
 |
Component:306; Jaccard:0.15404
 |
Component:306; Jaccard:0.12065
 |
Component:306; Jaccard:0.086877
 |
Correlation:0.549
 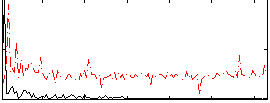 |
Correlation:0.549
  |
Correlation:0.549
  |
Correlation:0.549
  |
Correlation:0.549
  |
Correlation:0.549
  |
Correlation:0.549
  |
Component:089; Jaccard:0.096504
 |
Component:089; Jaccard:0.11244
 |
Component:241; Jaccard:0.12377
 |
Component:241; Jaccard:0.1353
 |
Component:241; Jaccard:0.13288
 |
Component:241; Jaccard:0.11552
 |
Component:241; Jaccard:0.085511
 |
Correlation:0.41382
 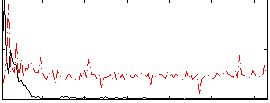 |
Correlation:0.41382
  |
Correlation:0.23847
  |
Correlation:0.23847
  |
Correlation:0.23847
  |
Correlation:0.23847
  |
Correlation:0.23847
  |
Component:219; Jaccard:0.090765
 |
Component:241; Jaccard:0.1069
 |
Component:089; Jaccard:0.1215
 |
Component:089; Jaccard:0.12146
 |
Component:251; Jaccard:0.1128
 |
Component:251; Jaccard:0.09117
 |
Component:251; Jaccard:0.069666
 |
Correlation:0.3198
  |
Correlation:0.23847
  |
Correlation:0.41382
  |
Correlation:0.41382
  |
Correlation:0.56895
  |
Correlation:0.56895
  |
Correlation:0.56895
  |
Component:117; Jaccard:0.089169
 |
Component:219; Jaccard:0.10558
 |
Component:251; Jaccard:0.11522
 |
Component:251; Jaccard:0.11969
 |
Component:219; Jaccard:0.10816
 |
Component:219; Jaccard:0.087251
 |
Component:219; Jaccard:0.062829
 |
Correlation:0.48393
  |
Correlation:0.3198
  |
Correlation:0.56895
  |
Correlation:0.56895
  |
Correlation:0.3198
  |
Correlation:0.3198
  |
Correlation:0.3198
  |
Component:234; Jaccard:0.088328
 |
Component:251; Jaccard:0.10288
 |
Component:219; Jaccard:0.11517
 |
Component:219; Jaccard:0.11856
 |
Component:089; Jaccard:0.10806
 |
Component:089; Jaccard:0.082265
 |
Component:089; Jaccard:0.058537
 |
Correlation:0.43377
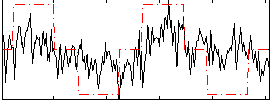  |
Correlation:0.56895
  |
Correlation:0.3198
  |
Correlation:0.3198
  |
Correlation:0.41382
  |
Correlation:0.41382
  |
Correlation:0.41382
  |
Component:251; Jaccard:0.088277
 |
Component:179; Jaccard:0.097887
 |
Component:179; Jaccard:0.10301
 |
Component:179; Jaccard:0.10263
 |
Component:179; Jaccard:0.091019
 |
Component:179; Jaccard:0.068318
 |
Component:031; Jaccard:0.045054
 |
Correlation:0.56895
  |
Correlation:0.47305
  |
Correlation:0.47305
  |
Correlation:0.47305
  |
Correlation:0.47305
  |
Correlation:0.47305
  |
Correlation:0.24946
  |
Component:241; Jaccard:0.087539
 |
Component:117; Jaccard:0.096049
 |
Component:234; Jaccard:0.093987
 |
Component:031; Jaccard:0.086899
 |
Component:031; Jaccard:0.078289
 |
Component:031; Jaccard:0.062115
 |
Component:179; Jaccard:0.044277
 |
Correlation:0.23847
  |
Correlation:0.48393
  |
Correlation:0.43377
  |
Correlation:0.24946
  |
Correlation:0.24946
  |
Correlation:0.24946
  |
Correlation:0.47305
  |
Component:179; Jaccard:0.086974
 |
Component:234; Jaccard:0.094916
 |
Component:117; Jaccard:0.093597
 |
Component:120; Jaccard:0.085485
 |
Component:120; Jaccard:0.069341
 |
Component:295; Jaccard:0.051777
 |
Component:295; Jaccard:0.036934
 |
Correlation:0.47305
  |
Correlation:0.43377
  |
Correlation:0.48393
  |
Correlation:0.32165
 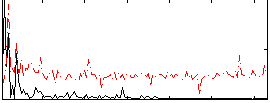 |
Correlation:0.32165
  |
Correlation:0.3217
  |
Correlation:0.3217
  |
Component:337; Jaccard:0.079909
 |
Component:120; Jaccard:0.087071
 |
Component:120; Jaccard:0.089902
 |
Component:117; Jaccard:0.085171
 |
Component:234; Jaccard:0.066812
 |
Component:234; Jaccard:0.050847
 |
Component:291; Jaccard:0.03465
 |
Correlation:0.51585
  |
Correlation:0.32165
  |
Correlation:0.32165
  |
Correlation:0.48393
  |
Correlation:0.43377
  |
Correlation:0.43377
  |
Correlation:0.34824
  |
Component:120; Jaccard:0.079475
 |
Component:337; Jaccard:0.08568
 |
Component:031; Jaccard:0.086798
 |
Component:234; Jaccard:0.084165
 |
Component:117; Jaccard:0.065268
 |
Component:120; Jaccard:0.046995
 |
Component:234; Jaccard:0.033228
 |
Correlation:0.32165
  |
Correlation:0.51585
  |
Correlation:0.24946
  |
Correlation:0.43377
  |
Correlation:0.48393
  |
Correlation:0.32165
  |
Correlation:0.43377
  |
Cope: 04:"neg_PUNISH" BACK
|
GLM result group-wise
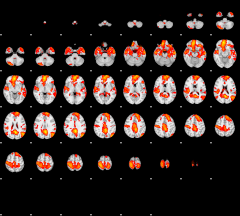 |
GLM binary result thresholded by 1
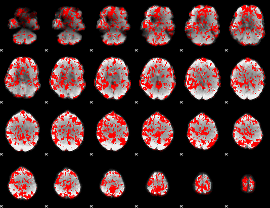 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
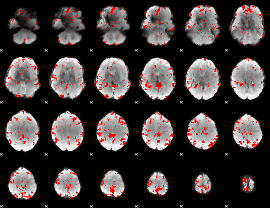 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
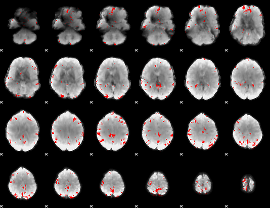 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:040; Jaccard:0.10172
 |
Component:344; Jaccard:0.1226
 |
Component:344; Jaccard:0.14729
 |
Component:344; Jaccard:0.16917
 |
Component:344; Jaccard:0.17512
 |
Component:344; Jaccard:0.16414
 |
Component:344; Jaccard:0.13143
 |
Correlation:0.13399
  |
Correlation:0.082305
  |
Correlation:0.082305
  |
Correlation:0.082305
  |
Correlation:0.082305
  |
Correlation:0.082305
  |
Correlation:0.082305
  |
Component:344; Jaccard:0.099108
 |
Component:392; Jaccard:0.11989
 |
Component:392; Jaccard:0.14224
 |
Component:392; Jaccard:0.1561
 |
Component:392; Jaccard:0.15676
 |
Component:392; Jaccard:0.13933
 |
Component:392; Jaccard:0.10811
 |
Correlation:0.082305
  |
Correlation:0.20281
  |
Correlation:0.20281
  |
Correlation:0.20281
  |
Correlation:0.20281
  |
Correlation:0.20281
  |
Correlation:0.20281
  |
Component:392; Jaccard:0.097923
 |
Component:396; Jaccard:0.11175
 |
Component:396; Jaccard:0.12305
 |
Component:396; Jaccard:0.12467
 |
Component:396; Jaccard:0.11546
 |
Component:396; Jaccard:0.096026
 |
Component:396; Jaccard:0.068051
 |
Correlation:0.20281
  |
Correlation:0.30129
 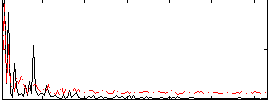 |
Correlation:0.30129
  |
Correlation:0.30129
  |
Correlation:0.30129
  |
Correlation:0.30129
  |
Correlation:0.30129
  |
Component:396; Jaccard:0.096986
 |
Component:040; Jaccard:0.10887
 |
Component:198; Jaccard:0.11784
 |
Component:198; Jaccard:0.11982
 |
Component:198; Jaccard:0.10881
 |
Component:198; Jaccard:0.085025
 |
Component:198; Jaccard:0.059865
 |
Correlation:0.30129
  |
Correlation:0.13399
  |
Correlation:0.30668
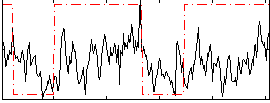 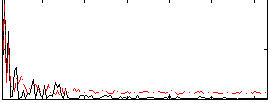 |
Correlation:0.30668
  |
Correlation:0.30668
  |
Correlation:0.30668
  |
Correlation:0.30668
  |
Component:198; Jaccard:0.094123
 |
Component:198; Jaccard:0.10847
 |
Component:040; Jaccard:0.11027
 |
Component:040; Jaccard:0.10338
 |
Component:040; Jaccard:0.087026
 |
Component:158; Jaccard:0.069848
 |
Component:086; Jaccard:0.055476
 |
Correlation:0.30668
  |
Correlation:0.30668
  |
Correlation:0.13399
  |
Correlation:0.13399
  |
Correlation:0.13399
  |
Correlation:0.094961
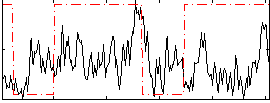  |
Correlation:0.24472
  |
Component:152; Jaccard:0.086478
 |
Component:088; Jaccard:0.093079
 |
Component:088; Jaccard:0.098108
 |
Component:088; Jaccard:0.099042
 |
Component:088; Jaccard:0.086935
 |
Component:088; Jaccard:0.069413
 |
Component:158; Jaccard:0.050535
 |
Correlation:0.41824
  |
Correlation:0.18056
  |
Correlation:0.18056
  |
Correlation:0.18056
  |
Correlation:0.18056
  |
Correlation:0.18056
  |
Correlation:0.094961
  |
Component:088; Jaccard:0.084126
 |
Component:361; Jaccard:0.09072
 |
Component:361; Jaccard:0.097688
 |
Component:361; Jaccard:0.097264
 |
Component:361; Jaccard:0.085898
 |
Component:086; Jaccard:0.069371
 |
Component:088; Jaccard:0.048666
 |
Correlation:0.18056
  |
Correlation:0.24467
  |
Correlation:0.24467
  |
Correlation:0.24467
  |
Correlation:0.24467
  |
Correlation:0.24472
  |
Correlation:0.18056
  |
Component:361; Jaccard:0.081826
 |
Component:152; Jaccard:0.089829
 |
Component:152; Jaccard:0.087234
 |
Component:158; Jaccard:0.087512
 |
Component:158; Jaccard:0.081745
 |
Component:361; Jaccard:0.065924
 |
Component:313; Jaccard:0.047342
 |
Correlation:0.24467
  |
Correlation:0.41824
  |
Correlation:0.41824
  |
Correlation:0.094961
  |
Correlation:0.094961
  |
Correlation:0.24467
  |
Correlation:-0.057965
  |
Component:103; Jaccard:0.078443
 |
Component:103; Jaccard:0.084151
 |
Component:103; Jaccard:0.087103
 |
Component:103; Jaccard:0.086004
 |
Component:086; Jaccard:0.081225
 |
Component:313; Jaccard:0.065518
 |
Component:040; Jaccard:0.045472
 |
Correlation:-0.088691
  |
Correlation:-0.088691
  |
Correlation:-0.088691
  |
Correlation:-0.088691
  |
Correlation:0.24472
  |
Correlation:-0.057965
  |
Correlation:0.13399
  |
Component:135; Jaccard:0.076798
 |
Component:135; Jaccard:0.083579
 |
Component:158; Jaccard:0.085806
 |
Component:086; Jaccard:0.084074
 |
Component:313; Jaccard:0.079559
 |
Component:040; Jaccard:0.065337
 |
Component:361; Jaccard:0.044297
 |
Correlation:-0.15622
  |
Correlation:-0.15622
  |
Correlation:0.094961
  |
Correlation:0.24472
  |
Correlation:-0.057965
  |
Correlation:0.13399
  |
Correlation:0.24467
  |
Cope: 05:"neg_REWARD" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
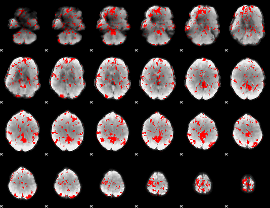 |
GLM binary result thresholded by 2.5
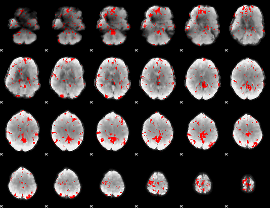 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
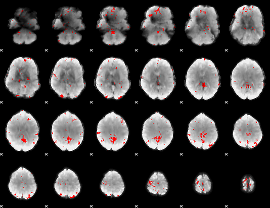 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
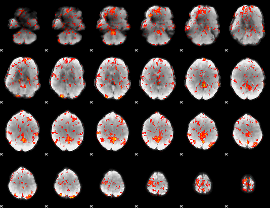 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
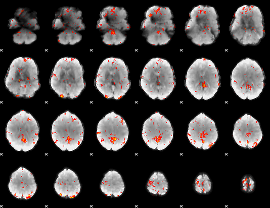 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:120; Jaccard:0.10082
 |
Component:120; Jaccard:0.12359
 |
Component:120; Jaccard:0.1504
 |
Component:120; Jaccard:0.17367
 |
Component:120; Jaccard:0.17887
 |
Component:120; Jaccard:0.1586
 |
Component:120; Jaccard:0.11846
 |
Correlation:0.44368
  |
Correlation:0.44368
  |
Correlation:0.44368
  |
Correlation:0.44368
  |
Correlation:0.44368
  |
Correlation:0.44368
  |
Correlation:0.44368
  |
Component:344; Jaccard:0.094657
 |
Component:344; Jaccard:0.1096
 |
Component:344; Jaccard:0.12424
 |
Component:344; Jaccard:0.13275
 |
Component:344; Jaccard:0.13273
 |
Component:344; Jaccard:0.12045
 |
Component:344; Jaccard:0.09468
 |
Correlation:0.27537
  |
Correlation:0.27537
  |
Correlation:0.27537
  |
Correlation:0.27537
  |
Correlation:0.27537
  |
Correlation:0.27537
  |
Correlation:0.27537
  |
Component:158; Jaccard:0.093129
 |
Component:158; Jaccard:0.10539
 |
Component:158; Jaccard:0.11726
 |
Component:158; Jaccard:0.12258
 |
Component:158; Jaccard:0.11374
 |
Component:158; Jaccard:0.093273
 |
Component:251; Jaccard:0.068016
 |
Correlation:0.20367
  |
Correlation:0.20367
  |
Correlation:0.20367
  |
Correlation:0.20367
  |
Correlation:0.20367
  |
Correlation:0.20367
  |
Correlation:0.54035
  |
Component:086; Jaccard:0.091147
 |
Component:086; Jaccard:0.10452
 |
Component:086; Jaccard:0.11351
 |
Component:086; Jaccard:0.11713
 |
Component:251; Jaccard:0.11014
 |
Component:251; Jaccard:0.092489
 |
Component:158; Jaccard:0.066844
 |
Correlation:0.18161
  |
Correlation:0.18161
  |
Correlation:0.18161
  |
Correlation:0.18161
  |
Correlation:0.54035
  |
Correlation:0.54035
  |
Correlation:0.20367
  |
Component:056; Jaccard:0.091003
 |
Component:056; Jaccard:0.10016
 |
Component:251; Jaccard:0.10973
 |
Component:251; Jaccard:0.11502
 |
Component:086; Jaccard:0.1088
 |
Component:086; Jaccard:0.089643
 |
Component:086; Jaccard:0.065795
 |
Correlation:0.46414
  |
Correlation:0.46414
  |
Correlation:0.54035
  |
Correlation:0.54035
  |
Correlation:0.18161
  |
Correlation:0.18161
  |
Correlation:0.18161
  |
Component:251; Jaccard:0.082771
 |
Component:251; Jaccard:0.097263
 |
Component:056; Jaccard:0.10547
 |
Component:056; Jaccard:0.10196
 |
Component:241; Jaccard:0.092892
 |
Component:241; Jaccard:0.075478
 |
Component:056; Jaccard:0.053753
 |
Correlation:0.54035
  |
Correlation:0.54035
  |
Correlation:0.46414
  |
Correlation:0.46414
  |
Correlation:0.23203
  |
Correlation:0.23203
  |
Correlation:0.46414
  |
Component:040; Jaccard:0.081478
 |
Component:306; Jaccard:0.091107
 |
Component:306; Jaccard:0.09734
 |
Component:241; Jaccard:0.10034
 |
Component:056; Jaccard:0.092168
 |
Component:056; Jaccard:0.075082
 |
Component:241; Jaccard:0.050388
 |
Correlation:0.088805
  |
Correlation:0.41201
  |
Correlation:0.41201
  |
Correlation:0.23203
  |
Correlation:0.46414
  |
Correlation:0.46414
  |
Correlation:0.23203
  |
Component:306; Jaccard:0.080821
 |
Component:326; Jaccard:0.085372
 |
Component:241; Jaccard:0.093394
 |
Component:306; Jaccard:0.094967
 |
Component:306; Jaccard:0.084998
 |
Component:306; Jaccard:0.066871
 |
Component:392; Jaccard:0.044246
 |
Correlation:0.41201
  |
Correlation:0.2432
  |
Correlation:0.23203
  |
Correlation:0.41201
  |
Correlation:0.41201
  |
Correlation:0.41201
  |
Correlation:0.16763
  |
Component:326; Jaccard:0.078853
 |
Component:241; Jaccard:0.082461
 |
Component:326; Jaccard:0.088744
 |
Component:089; Jaccard:0.085597
 |
Component:089; Jaccard:0.073373
 |
Component:392; Jaccard:0.060028
 |
Component:306; Jaccard:0.043538
 |
Correlation:0.2432
  |
Correlation:0.23203
  |
Correlation:0.2432
  |
Correlation:0.29251
  |
Correlation:0.29251
  |
Correlation:0.16763
  |
Correlation:0.41201
  |
Component:268; Jaccard:0.07684
 |
Component:040; Jaccard:0.082457
 |
Component:089; Jaccard:0.087649
 |
Component:326; Jaccard:0.08286
 |
Component:326; Jaccard:0.071267
 |
Component:089; Jaccard:0.057808
 |
Component:219; Jaccard:0.043166
 |
Correlation:0.15321
  |
Correlation:0.088805
  |
Correlation:0.29251
  |
Correlation:0.2432
  |
Correlation:0.2432
  |
Correlation:0.29251
  |
Correlation:0.25903
  |
Cope: 06:"REWARD-PUNISH" BACK
|
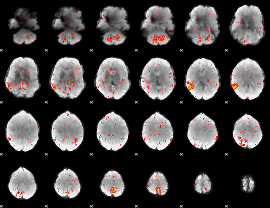
GLM result group-wise
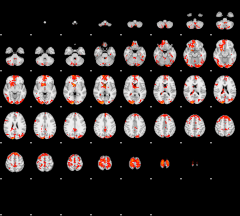 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
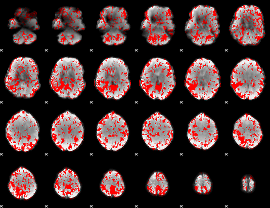 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
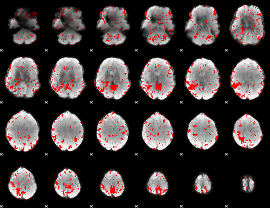 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
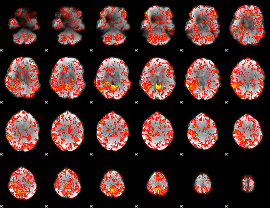 |
GLM Z-score result thresholded by 1.5
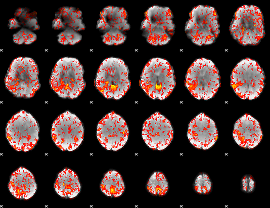 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
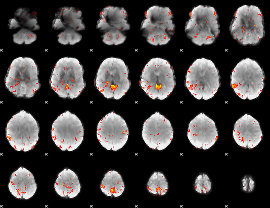 |
GLM Z-score result thresholded by 4
 |
Component:377; Jaccard:0.10763
 |
Component:377; Jaccard:0.12276
 |
Component:181; Jaccard:0.1348
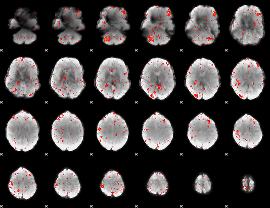 |
Component:181; Jaccard:0.14139
 |
Component:034; Jaccard:0.15669
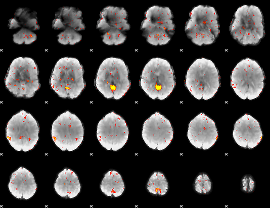 |
Component:034; Jaccard:0.16905
 |
Component:034; Jaccard:0.16859
 |
Correlation:0.25734
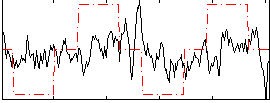 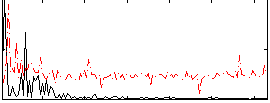 |
Correlation:0.25734
  |
Correlation:0.50305
  |
Correlation:0.50305
  |
Correlation:0.5236
  |
Correlation:0.5236
  |
Correlation:0.5236
  |
Component:181; Jaccard:0.099525
 |
Component:181; Jaccard:0.11927
 |
Component:377; Jaccard:0.13206
 |
Component:034; Jaccard:0.133
 |
Component:260; Jaccard:0.14443
 |
Component:260; Jaccard:0.14413
 |
Component:260; Jaccard:0.13078
 |
Correlation:0.50305
  |
Correlation:0.50305
  |
Correlation:0.25734
  |
Correlation:0.5236
  |
Correlation:0.4433
  |
Correlation:0.4433
  |
Correlation:0.4433
  |
Component:236; Jaccard:0.077628
 |
Component:236; Jaccard:0.095938
 |
Component:236; Jaccard:0.11397
 |
Component:377; Jaccard:0.13226
 |
Component:236; Jaccard:0.13721
 |
Component:236; Jaccard:0.12549
 |
Component:236; Jaccard:0.10386
 |
Correlation:0.22994
  |
Correlation:0.22994
  |
Correlation:0.22994
  |
Correlation:0.25734
  |
Correlation:0.22994
  |
Correlation:0.22994
  |
Correlation:0.22994
  |
Component:329; Jaccard:0.077032
 |
Component:260; Jaccard:0.088431
 |
Component:260; Jaccard:0.10844
 |
Component:236; Jaccard:0.13096
 |
Component:181; Jaccard:0.13631
 |
Component:181; Jaccard:0.11938
 |
Component:181; Jaccard:0.091596
 |
Correlation:-0.036084
  |
Correlation:0.4433
  |
Correlation:0.4433
  |
Correlation:0.22994
  |
Correlation:0.50305
  |
Correlation:0.50305
  |
Correlation:0.50305
  |
Component:115; Jaccard:0.07501
 |
Component:034; Jaccard:0.085559
 |
Component:034; Jaccard:0.10762
 |
Component:260; Jaccard:0.1288
 |
Component:377; Jaccard:0.11776
 |
Component:206; Jaccard:0.093683
 |
Component:206; Jaccard:0.082423
 |
Correlation:-0.19916
  |
Correlation:0.5236
  |
Correlation:0.5236
  |
Correlation:0.4433
  |
Correlation:0.25734
  |
Correlation:0.60181
  |
Correlation:0.60181
  |
Component:361; Jaccard:0.073132
 |
Component:163; Jaccard:0.08059
 |
Component:163; Jaccard:0.088204
 |
Component:163; Jaccard:0.091325
 |
Component:206; Jaccard:0.093154
 |
Component:377; Jaccard:0.090963
 |
Component:377; Jaccard:0.062243
 |
Correlation:0.25511
  |
Correlation:0.27835
  |
Correlation:0.27835
  |
Correlation:0.27835
  |
Correlation:0.60181
  |
Correlation:0.25734
  |
Correlation:0.25734
  |
Component:135; Jaccard:0.07173
 |
Component:361; Jaccard:0.080106
 |
Component:332; Jaccard:0.084768
 |
Component:332; Jaccard:0.089832
 |
Component:163; Jaccard:0.088518
 |
Component:332; Jaccard:0.076532
 |
Component:332; Jaccard:0.061912
 |
Correlation:-0.22274
  |
Correlation:0.25511
  |
Correlation:0.5003
  |
Correlation:0.5003
  |
Correlation:0.27835
  |
Correlation:0.5003
  |
Correlation:0.5003
  |
Component:393; Jaccard:0.071453
 |
Component:329; Jaccard:0.077933
 |
Component:361; Jaccard:0.082664
 |
Component:206; Jaccard:0.085891
 |
Component:332; Jaccard:0.085289
 |
Component:163; Jaccard:0.075011
 |
Component:002; Jaccard:0.058915
 |
Correlation:-0.02005
  |
Correlation:-0.036084
  |
Correlation:0.25511
  |
Correlation:0.60181
  |
Correlation:0.5003
  |
Correlation:0.27835
  |
Correlation:0.25537
  |
Component:260; Jaccard:0.071223
 |
Component:393; Jaccard:0.077099
 |
Component:393; Jaccard:0.078498
 |
Component:361; Jaccard:0.079976
 |
Component:002; Jaccard:0.080983
 |
Component:002; Jaccard:0.074382
 |
Component:163; Jaccard:0.056188
 |
Correlation:0.4433
  |
Correlation:-0.02005
  |
Correlation:-0.02005
  |
Correlation:0.25511
  |
Correlation:0.25537
  |
Correlation:0.25537
  |
Correlation:0.27835
  |
Component:152; Jaccard:0.070836
 |
Component:115; Jaccard:0.07637
 |
Component:178; Jaccard:0.077578
 |
Component:002; Jaccard:0.079518
 |
Component:020; Jaccard:0.07808
 |
Component:020; Jaccard:0.06692
 |
Component:020; Jaccard:0.052174
 |
Correlation:0.36947
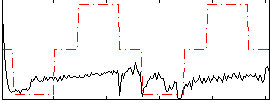  |
Correlation:-0.19916
  |
Correlation:0.33748
 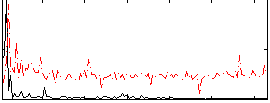 |
Correlation:0.25537
  |
Correlation:0.27833
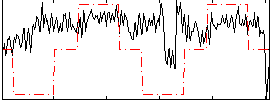  |
Correlation:0.27833
  |
Correlation:0.27833
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































