Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
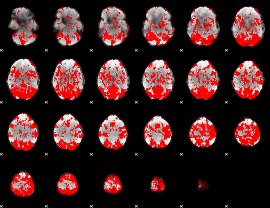 |
GLM binary result thresholded by 1.5
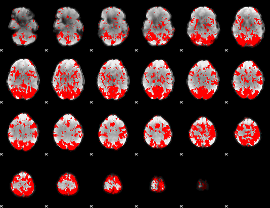 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
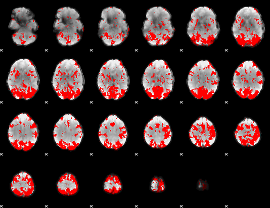
GLM binary result thresholded by 3
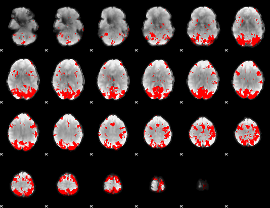 |
GLM binary result thresholded by 3.5
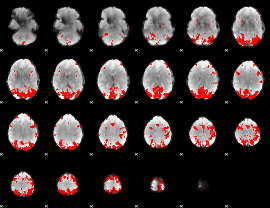 |
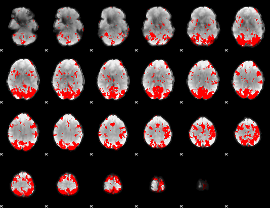
GLM binary result thresholded by 4
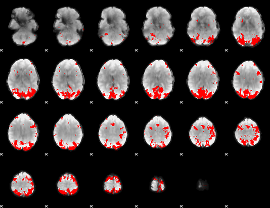 |
GLM Z-score result thresholded by 1
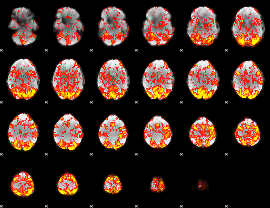 |
GLM Z-score result thresholded by 1.5
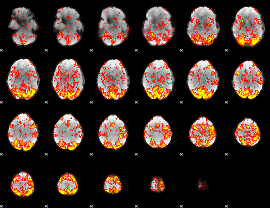 |
GLM Z-score result thresholded by 2
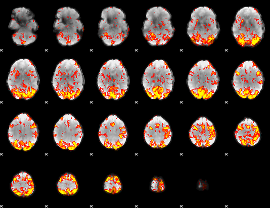 |
GLM Z-score result thresholded by 2.5
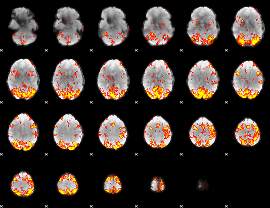 |
GLM Z-score result thresholded by 3
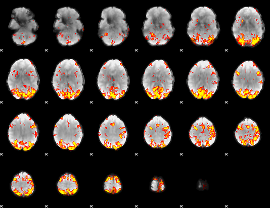 |
GLM Z-score result thresholded by 3.5
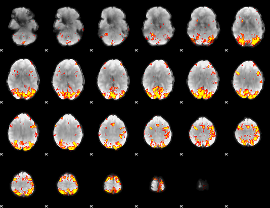 |
GLM Z-score result thresholded by 4
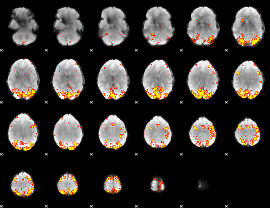 |
Component:199; Jaccard:0.1583
 |
Component:199; Jaccard:0.19577
 |
Component:199; Jaccard:0.23852
 |
Component:199; Jaccard:0.28219
 |
Component:146; Jaccard:0.3283
 |
Component:146; Jaccard:0.37123
 |
Component:146; Jaccard:0.40455
 |
Correlation:0.10945
  |
Correlation:0.10945
  |
Correlation:0.10945
  |
Correlation:0.10945
  |
Correlation:0.24099
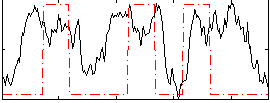 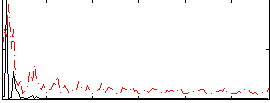 |
Correlation:0.24099
  |
Correlation:0.24099
  |
Component:146; Jaccard:0.14237
 |
Component:146; Jaccard:0.18007
 |
Component:146; Jaccard:0.22566
 |
Component:146; Jaccard:0.27703
 |
Component:199; Jaccard:0.3218
 |
Component:199; Jaccard:0.35089
 |
Component:199; Jaccard:0.36673
 |
Correlation:0.24099
  |
Correlation:0.24099
  |
Correlation:0.24099
  |
Correlation:0.24099
  |
Correlation:0.10945
  |
Correlation:0.10945
  |
Correlation:0.10945
  |
Component:384; Jaccard:0.12586
 |
Component:384; Jaccard:0.14967
 |
Component:384; Jaccard:0.17391
 |
Component:214; Jaccard:0.2041
 |
Component:214; Jaccard:0.23287
 |
Component:214; Jaccard:0.25577
 |
Component:214; Jaccard:0.27177
 |
Correlation:0.11589
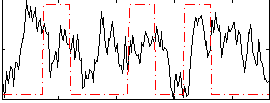 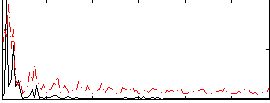 |
Correlation:0.11589
  |
Correlation:0.11589
  |
Correlation:0.089344
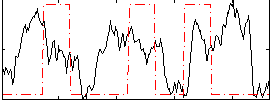 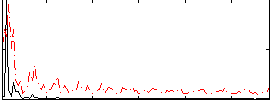 |
Correlation:0.089344
  |
Correlation:0.089344
  |
Correlation:0.089344
  |
Component:062; Jaccard:0.12247
 |
Component:062; Jaccard:0.14555
 |
Component:214; Jaccard:0.17319
 |
Component:385; Jaccard:0.19873
 |
Component:385; Jaccard:0.2218
 |
Component:385; Jaccard:0.23995
 |
Component:385; Jaccard:0.24827
 |
Correlation:0.32358
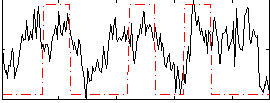 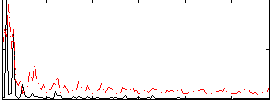 |
Correlation:0.32358
  |
Correlation:0.089344
  |
Correlation:0.084609
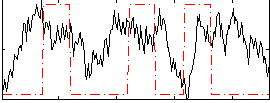 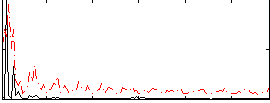 |
Correlation:0.084609
  |
Correlation:0.084609
  |
Correlation:0.084609
  |
Component:127; Jaccard:0.11956
 |
Component:214; Jaccard:0.14428
 |
Component:385; Jaccard:0.1704
 |
Component:384; Jaccard:0.19568
 |
Component:384; Jaccard:0.20866
 |
Component:384; Jaccard:0.21214
 |
Component:184; Jaccard:0.21537
 |
Correlation:0.10918
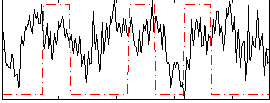 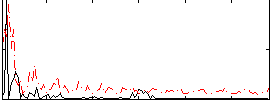 |
Correlation:0.089344
  |
Correlation:0.084609
  |
Correlation:0.11589
  |
Correlation:0.11589
  |
Correlation:0.11589
  |
Correlation:0.15731
  |
Component:214; Jaccard:0.11848
 |
Component:385; Jaccard:0.1412
 |
Component:062; Jaccard:0.16868
 |
Component:062; Jaccard:0.1871
 |
Component:062; Jaccard:0.20098
 |
Component:184; Jaccard:0.21148
 |
Component:384; Jaccard:0.21321
 |
Correlation:0.089344
  |
Correlation:0.084609
  |
Correlation:0.32358
  |
Correlation:0.32358
  |
Correlation:0.32358
  |
Correlation:0.15731
  |
Correlation:0.11589
  |
Component:184; Jaccard:0.11643
 |
Component:127; Jaccard:0.13921
 |
Component:184; Jaccard:0.16281
 |
Component:184; Jaccard:0.18325
 |
Component:184; Jaccard:0.20048
 |
Component:062; Jaccard:0.20877
 |
Component:062; Jaccard:0.21134
 |
Correlation:0.15731
  |
Correlation:0.10918
  |
Correlation:0.15731
  |
Correlation:0.15731
  |
Correlation:0.15731
  |
Correlation:0.32358
  |
Correlation:0.32358
  |
Component:123; Jaccard:0.11509
 |
Component:184; Jaccard:0.13917
 |
Component:127; Jaccard:0.15875
 |
Component:127; Jaccard:0.17454
 |
Component:127; Jaccard:0.1846
 |
Component:027; Jaccard:0.19076
 |
Component:027; Jaccard:0.19747
 |
Correlation:0.18386
 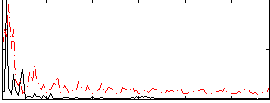 |
Correlation:0.15731
  |
Correlation:0.10918
  |
Correlation:0.10918
  |
Correlation:0.10918
  |
Correlation:0.15534
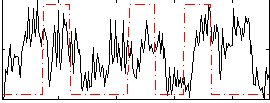 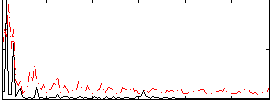 |
Correlation:0.15534
  |
Component:385; Jaccard:0.11497
 |
Component:294; Jaccard:0.13305
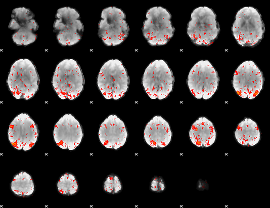 |
Component:294; Jaccard:0.15416
 |
Component:294; Jaccard:0.17095
 |
Component:294; Jaccard:0.18314
 |
Component:123; Jaccard:0.19047
 |
Component:123; Jaccard:0.19108
 |
Correlation:0.084609
  |
Correlation:0.19053
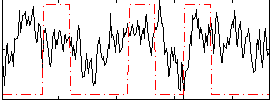 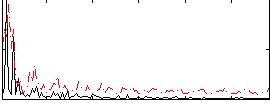 |
Correlation:0.19053
  |
Correlation:0.19053
  |
Correlation:0.19053
  |
Correlation:0.18386
  |
Correlation:0.18386
  |
Component:260; Jaccard:0.11371
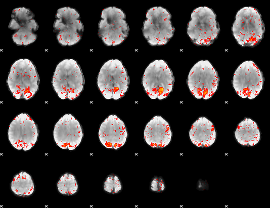 |
Component:123; Jaccard:0.13287
 |
Component:123; Jaccard:0.15136
 |
Component:123; Jaccard:0.16769
 |
Component:123; Jaccard:0.18217
 |
Component:127; Jaccard:0.18726
 |
Component:294; Jaccard:0.18437
 |
Correlation:0.15792
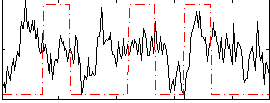 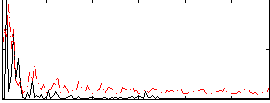 |
Correlation:0.18386
  |
Correlation:0.18386
  |
Correlation:0.18386
  |
Correlation:0.18386
  |
Correlation:0.10918
  |
Correlation:0.19053
  |
Cope: 02:"REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
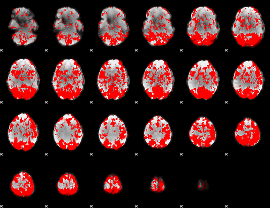 |
GLM binary result thresholded by 1.5
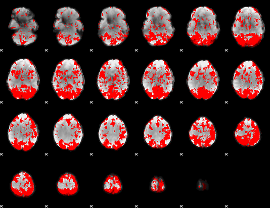 |
GLM binary result thresholded by 2
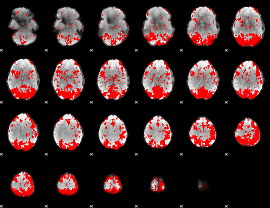 |
GLM binary result thresholded by 2.5
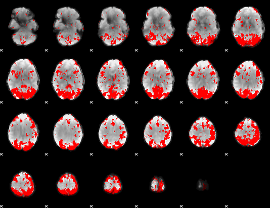 |
GLM binary result thresholded by 3
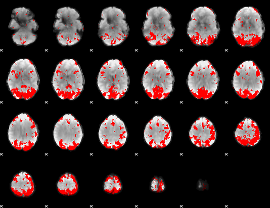 |
GLM binary result thresholded by 3.5
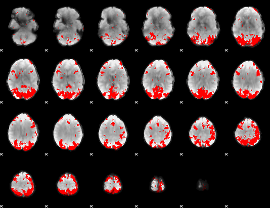 |
GLM binary result thresholded by 4
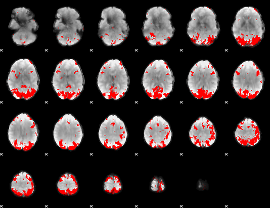 |
GLM Z-score result thresholded by 1
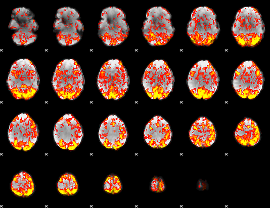 |
GLM Z-score result thresholded by 1.5
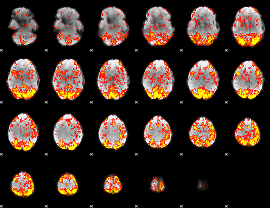 |
GLM Z-score result thresholded by 2
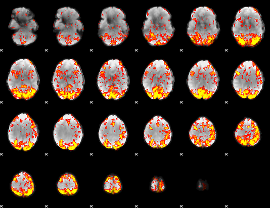 |
GLM Z-score result thresholded by 2.5
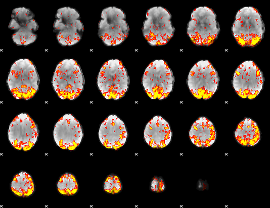 |
GLM Z-score result thresholded by 3
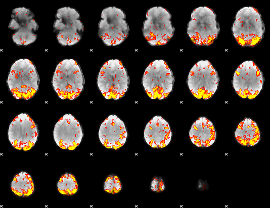 |
GLM Z-score result thresholded by 3.5
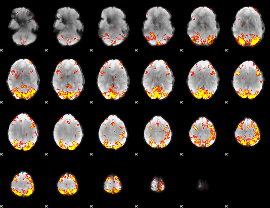 |
GLM Z-score result thresholded by 4
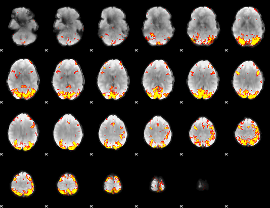 |
Component:199; Jaccard:0.15275
 |
Component:199; Jaccard:0.18434
 |
Component:199; Jaccard:0.22007
 |
Component:199; Jaccard:0.25892
 |
Component:146; Jaccard:0.30366
 |
Component:146; Jaccard:0.35229
 |
Component:146; Jaccard:0.39694
 |
Correlation:0.35203
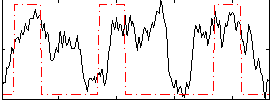  |
Correlation:0.35203
  |
Correlation:0.35203
  |
Correlation:0.35203
  |
Correlation:0.35735
  |
Correlation:0.35735
  |
Correlation:0.35735
  |
Component:146; Jaccard:0.13832
 |
Component:146; Jaccard:0.1701
 |
Component:146; Jaccard:0.20876
 |
Component:146; Jaccard:0.25416
 |
Component:199; Jaccard:0.29733
 |
Component:199; Jaccard:0.32666
 |
Component:199; Jaccard:0.35278
 |
Correlation:0.35735
  |
Correlation:0.35735
  |
Correlation:0.35735
  |
Correlation:0.35735
  |
Correlation:0.35203
  |
Correlation:0.35203
  |
Correlation:0.35203
  |
Component:214; Jaccard:0.12265
 |
Component:214; Jaccard:0.14918
 |
Component:214; Jaccard:0.18048
 |
Component:214; Jaccard:0.2143
 |
Component:214; Jaccard:0.24729
 |
Component:214; Jaccard:0.27965
 |
Component:214; Jaccard:0.30666
 |
Correlation:0.54147
  |
Correlation:0.54147
  |
Correlation:0.54147
  |
Correlation:0.54147
  |
Correlation:0.54147
  |
Correlation:0.54147
  |
Correlation:0.54147
  |
Component:127; Jaccard:0.12086
 |
Component:127; Jaccard:0.13934
 |
Component:385; Jaccard:0.1603
 |
Component:385; Jaccard:0.18769
 |
Component:385; Jaccard:0.21362
 |
Component:385; Jaccard:0.23653
 |
Component:385; Jaccard:0.25406
 |
Correlation:0.25937
  |
Correlation:0.25937
  |
Correlation:0.33508
  |
Correlation:0.33508
  |
Correlation:0.33508
  |
Correlation:0.33508
  |
Correlation:0.33508
  |
Component:384; Jaccard:0.11836
 |
Component:384; Jaccard:0.13642
 |
Component:127; Jaccard:0.15784
 |
Component:184; Jaccard:0.17693
 |
Component:184; Jaccard:0.19596
 |
Component:184; Jaccard:0.21018
 |
Component:184; Jaccard:0.21888
 |
Correlation:0.40243
  |
Correlation:0.40243
  |
Correlation:0.25937
  |
Correlation:0.40598
 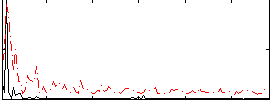 |
Correlation:0.40598
  |
Correlation:0.40598
  |
Correlation:0.40598
  |
Component:062; Jaccard:0.11541
 |
Component:385; Jaccard:0.1352
 |
Component:184; Jaccard:0.15631
 |
Component:127; Jaccard:0.1754
 |
Component:127; Jaccard:0.18839
 |
Component:127; Jaccard:0.19631
 |
Component:062; Jaccard:0.19972
 |
Correlation:0.1643
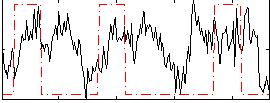 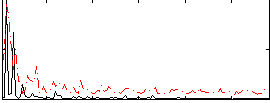 |
Correlation:0.33508
  |
Correlation:0.40598
  |
Correlation:0.25937
  |
Correlation:0.25937
  |
Correlation:0.25937
  |
Correlation:0.1643
  |
Component:184; Jaccard:0.11463
 |
Component:184; Jaccard:0.13485
 |
Component:384; Jaccard:0.15465
 |
Component:384; Jaccard:0.17029
 |
Component:384; Jaccard:0.1844
 |
Component:062; Jaccard:0.19404
 |
Component:127; Jaccard:0.19852
 |
Correlation:0.40598
  |
Correlation:0.40598
  |
Correlation:0.40243
  |
Correlation:0.40243
  |
Correlation:0.40243
  |
Correlation:0.1643
  |
Correlation:0.25937
  |
Component:385; Jaccard:0.11265
 |
Component:062; Jaccard:0.13317
 |
Component:062; Jaccard:0.15164
 |
Component:062; Jaccard:0.16856
 |
Component:062; Jaccard:0.18393
 |
Component:384; Jaccard:0.19314
 |
Component:384; Jaccard:0.19587
 |
Correlation:0.33508
  |
Correlation:0.1643
  |
Correlation:0.1643
  |
Correlation:0.1643
  |
Correlation:0.1643
  |
Correlation:0.40243
  |
Correlation:0.40243
  |
Component:123; Jaccard:0.10688
 |
Component:123; Jaccard:0.12001
 |
Component:123; Jaccard:0.13373
 |
Component:123; Jaccard:0.14739
 |
Component:123; Jaccard:0.15929
 |
Component:123; Jaccard:0.16758
 |
Component:123; Jaccard:0.17305
 |
Correlation:0.2665
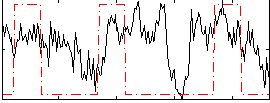  |
Correlation:0.2665
  |
Correlation:0.2665
  |
Correlation:0.2665
  |
Correlation:0.2665
  |
Correlation:0.2665
  |
Correlation:0.2665
  |
Component:260; Jaccard:0.10142
 |
Component:294; Jaccard:0.11497
 |
Component:294; Jaccard:0.12857
 |
Component:294; Jaccard:0.14168
 |
Component:294; Jaccard:0.15206
 |
Component:027; Jaccard:0.16235
 |
Component:027; Jaccard:0.17049
 |
Correlation:0.36523
  |
Correlation:0.26911
  |
Correlation:0.26911
  |
Correlation:0.26911
  |
Correlation:0.26911
  |
Correlation:0.24522
  |
Correlation:0.24522
  |
Cope: 03:"MATCH-REL" BACK
|
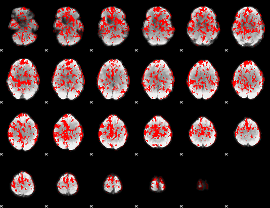
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
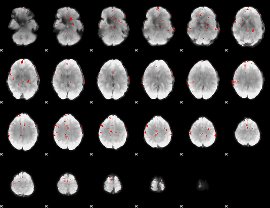 |
GLM binary result thresholded by 4
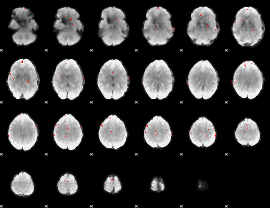 |
GLM Z-score result thresholded by 1
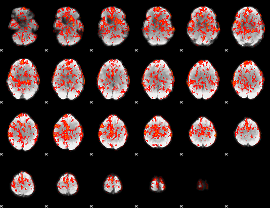 |
GLM Z-score result thresholded by 1.5
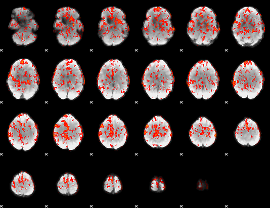 |
GLM Z-score result thresholded by 2
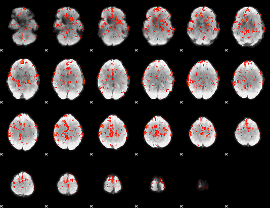 |
GLM Z-score result thresholded by 2.5
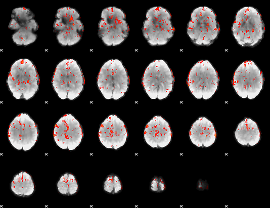 |
GLM Z-score result thresholded by 3
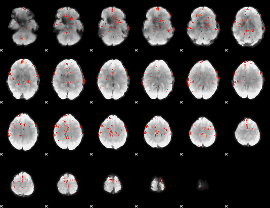 |
GLM Z-score result thresholded by 3.5
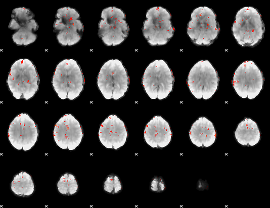 |
GLM Z-score result thresholded by 4
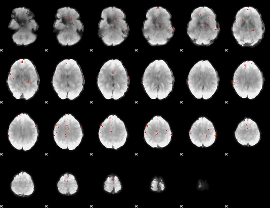 |
Component:281; Jaccard:0.10491
 |
Component:262; Jaccard:0.12746
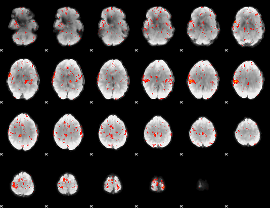
 |
Component:262; Jaccard:0.16024
 |
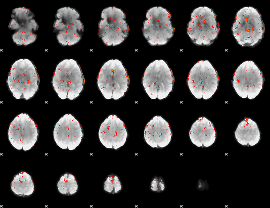
Component:262; Jaccard:0.18031
 |
Component:262; Jaccard:0.17019
 |
Component:262; Jaccard:0.11737
 |
Component:262; Jaccard:0.060047
 |
Correlation:0.32112
  |
Correlation:0.071291
  |
Correlation:0.071291
  |
Correlation:0.071291
  |
Correlation:0.071291
  |
Correlation:0.071291
  |
Correlation:0.071291
  |
Component:262; Jaccard:0.098834
 |
Component:281; Jaccard:0.12215
 |
Component:281; Jaccard:0.13321
 |
Component:281; Jaccard:0.12773
 |
Component:281; Jaccard:0.10291
 |
Component:278; Jaccard:0.062172
 |
Component:278; Jaccard:0.035314
 |
Correlation:0.071291
  |
Correlation:0.32112
  |
Correlation:0.32112
  |
Correlation:0.32112
  |
Correlation:0.32112
  |
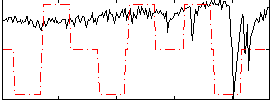
Correlation:0.30722
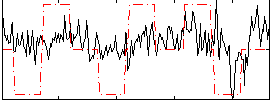  |
Correlation:0.30722
  |
Component:278; Jaccard:0.090511
 |
Component:278; Jaccard:0.10552
 |
Component:278; Jaccard:0.11419
 |
Component:278; Jaccard:0.11451
 |
Component:278; Jaccard:0.095215
 |
Component:281; Jaccard:0.060223
 |
Component:371; Jaccard:0.029784
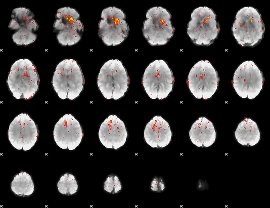 |
Correlation:0.30722
  |
Correlation:0.30722
  |
Correlation:0.30722
  |
Correlation:0.30722
  |
Correlation:0.30722
  |
Correlation:0.32112
  |
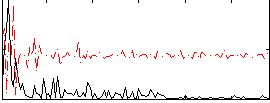
Correlation:0.17827
 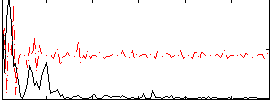 |
Component:399; Jaccard:0.084681
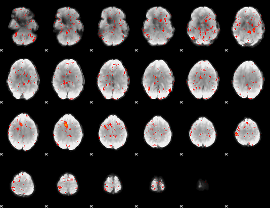 |
Component:399; Jaccard:0.091872
 |
Component:371; Jaccard:0.10414
 |
Component:371; Jaccard:0.10383
 |
Component:371; Jaccard:0.089794
 |
Component:371; Jaccard:0.059652
 |
Component:116; Jaccard:0.029282
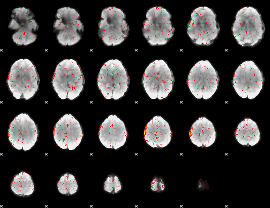 |
Correlation:0.17162
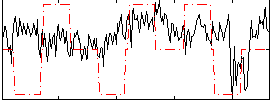 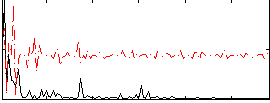 |
Correlation:0.17162
  |
Correlation:0.17827
  |
Correlation:0.17827
  |
Correlation:0.17827
  |
Correlation:0.17827
  |
Correlation:0.24591
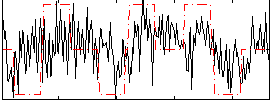 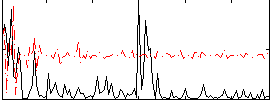 |
Component:308; Jaccard:0.080329
 |
Component:371; Jaccard:0.09027
 |
Component:133; Jaccard:0.093642
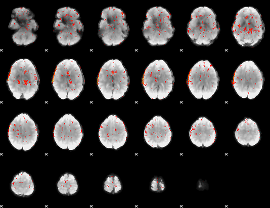 |
Component:133; Jaccard:0.095534
 |
Component:133; Jaccard:0.083103
 |
Component:103; Jaccard:0.05094
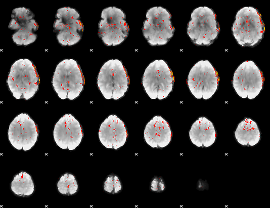 |
Component:281; Jaccard:0.02914
 |
Correlation:0.13265
  |
Correlation:0.17827
  |
Correlation:0.22206
 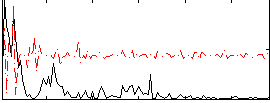 |
Correlation:0.22206
  |
Correlation:0.22206
  |
Correlation:0.043018
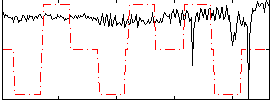 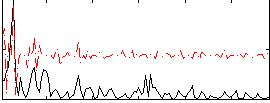 |
Correlation:0.32112
  |
Component:356; Jaccard:0.07662
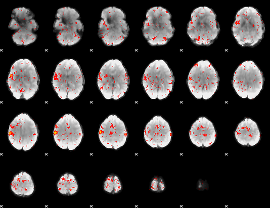 |
Component:308; Jaccard:0.085372
 |
Component:399; Jaccard:0.093304
 |
Component:103; Jaccard:0.090363
 |
Component:103; Jaccard:0.078392
 |
Component:133; Jaccard:0.050182
 |
Component:256; Jaccard:0.026952
 |
Correlation:0.011244
  |
Correlation:0.13265
  |
Correlation:0.17162
  |
Correlation:0.043018
  |
Correlation:0.043018
  |
Correlation:0.22206
  |
Correlation:0.19299
  |
Component:371; Jaccard:0.075029
 |
Component:133; Jaccard:0.082105
 |
Component:103; Jaccard:0.088355
 |
Component:256; Jaccard:0.083576
 |
Component:293; Jaccard:0.068755
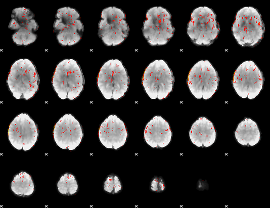 |
Component:256; Jaccard:0.048136
 |
Component:133; Jaccard:0.025182
 |
Correlation:0.17827
  |
Correlation:0.22206
  |
Correlation:0.043018
  |
Correlation:0.19299
  |
Correlation:0.32726
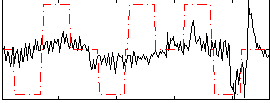 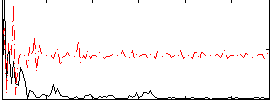 |
Correlation:0.19299
  |
Correlation:0.22206
  |
Component:042; Jaccard:0.074626
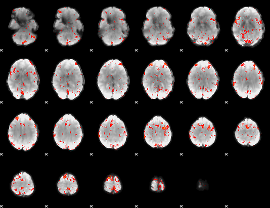 |
Component:256; Jaccard:0.081314
 |
Component:293; Jaccard:0.085743
 |
Component:293; Jaccard:0.082771
 |
Component:256; Jaccard:0.067473
 |
Component:116; Jaccard:0.048108
 |
Component:103; Jaccard:0.024731
 |
Correlation:0.34462
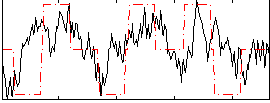 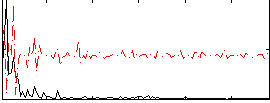 |
Correlation:0.19299
  |
Correlation:0.32726
  |
Correlation:0.32726
  |
Correlation:0.19299
  |
Correlation:0.24591
  |
Correlation:0.043018
  |
Component:311; Jaccard:0.073297
 |
Component:356; Jaccard:0.080424
 |
Component:308; Jaccard:0.085101
 |
Component:399; Jaccard:0.082094
 |
Component:399; Jaccard:0.063304
 |
Component:399; Jaccard:0.042305
 |
Component:399; Jaccard:0.022304
 |
Correlation:0.18844
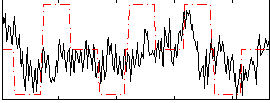 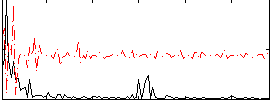 |
Correlation:0.011244
  |
Correlation:0.13265
  |
Correlation:0.17162
  |
Correlation:0.17162
  |
Correlation:0.17162
  |
Correlation:0.17162
  |
Component:256; Jaccard:0.07248
 |
Component:311; Jaccard:0.07904
 |
Component:256; Jaccard:0.084938
 |
Component:308; Jaccard:0.076337
 |
Component:116; Jaccard:0.062987
 |
Component:293; Jaccard:0.040935
 |
Component:356; Jaccard:0.020808
 |
Correlation:0.19299
  |
Correlation:0.18844
  |
Correlation:0.19299
  |
Correlation:0.13265
  |
Correlation:0.24591
  |
Correlation:0.32726
  |
Correlation:0.011244
  |
Cope: 04:"REL-MATCH" BACK
|
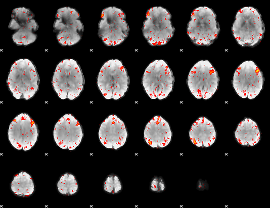
GLM result group-wise
 |
GLM binary result thresholded by 1
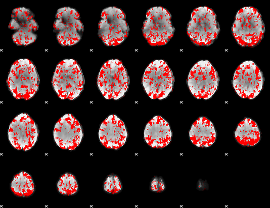 |
GLM binary result thresholded by 1.5
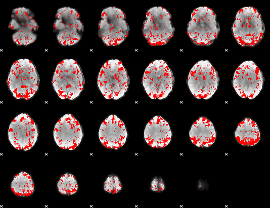 |
GLM binary result thresholded by 2
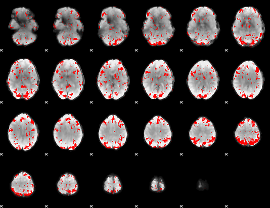 |
GLM binary result thresholded by 2.5
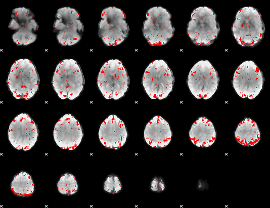 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
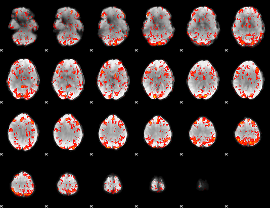 |
GLM Z-score result thresholded by 2
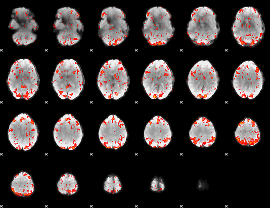 |
GLM Z-score result thresholded by 2.5
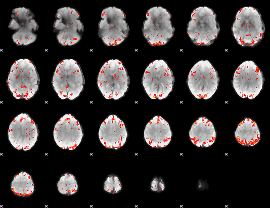 |
GLM Z-score result thresholded by 3
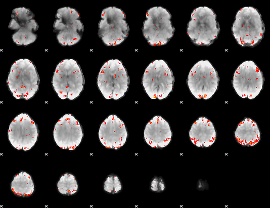 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:214; Jaccard:0.12688
 |
Component:264; Jaccard:0.14775
 |
Component:264; Jaccard:0.16445
 |
Component:264; Jaccard:0.1691
 |
Component:264; Jaccard:0.1468
 |
Component:264; Jaccard:0.10803
 |
Component:264; Jaccard:0.066054
 |
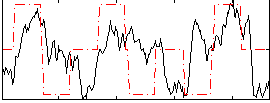
Correlation:0.26797
 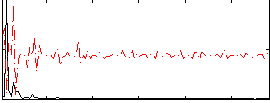 |
Correlation:0.15389
 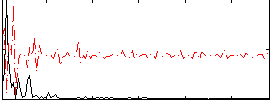 |
Correlation:0.15389
  |
Correlation:0.15389
  |
Correlation:0.15389
  |
Correlation:0.15389
  |
Correlation:0.15389
  |
Component:264; Jaccard:0.12296
 |
Component:214; Jaccard:0.14513
 |
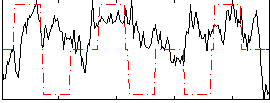
Component:155; Jaccard:0.16047
 |
Component:155; Jaccard:0.1654
 |
Component:155; Jaccard:0.14597
 |
Component:155; Jaccard:0.10691
 |
Component:155; Jaccard:0.065286
 |
Correlation:0.15389
  |
Correlation:0.26797
  |
Correlation:0.39337
  |
Correlation:0.39337
  |
Correlation:0.39337
  |
Correlation:0.39337
  |
Correlation:0.39337
  |
Component:155; Jaccard:0.12102
 |
Component:155; Jaccard:0.14351
 |
Component:214; Jaccard:0.15603
 |
Component:289; Jaccard:0.15294
 |
Component:289; Jaccard:0.13366
 |
Component:289; Jaccard:0.099279
 |
Component:289; Jaccard:0.064694
 |
Correlation:0.39337
  |
Correlation:0.39337
  |
Correlation:0.26797
  |
Correlation:0.32179
 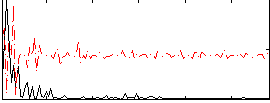 |
Correlation:0.32179
  |
Correlation:0.32179
  |
Correlation:0.32179
  |
Component:146; Jaccard:0.11682
 |
Component:289; Jaccard:0.13448
 |
Component:289; Jaccard:0.14866
 |
Component:214; Jaccard:0.14744
 |
Component:214; Jaccard:0.12275
 |
Component:214; Jaccard:0.087555
 |
Component:214; Jaccard:0.053745
 |
Correlation:0.068967
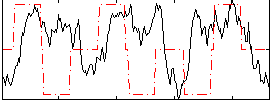 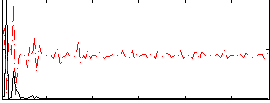 |
Correlation:0.32179
  |
Correlation:0.32179
  |
Correlation:0.26797
  |
Correlation:0.26797
  |
Correlation:0.26797
  |
Correlation:0.26797
  |
Component:289; Jaccard:0.11308
 |
Component:146; Jaccard:0.12924
 |
Component:146; Jaccard:0.13322
 |
Component:313; Jaccard:0.12131
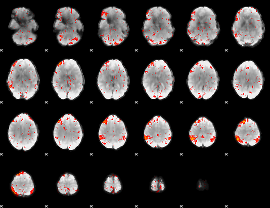 |
Component:313; Jaccard:0.10558
 |
Component:313; Jaccard:0.077822
 |
Component:250; Jaccard:0.049489
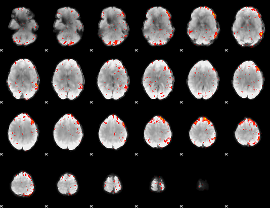 |
Correlation:0.32179
  |
Correlation:0.068967
  |
Correlation:0.068967
  |
Correlation:0.11157
  |
Correlation:0.11157
  |
Correlation:0.11157
  |
Correlation:0.38158
 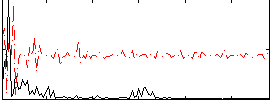 |
Component:199; Jaccard:0.105
 |
Component:313; Jaccard:0.11399
 |
Component:313; Jaccard:0.12173
 |
Component:146; Jaccard:0.11994
 |
Component:250; Jaccard:0.096284
 |
Component:250; Jaccard:0.074062
 |
Component:313; Jaccard:0.048424
 |
Correlation:0.14377
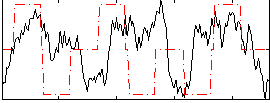 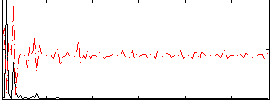 |
Correlation:0.11157
  |
Correlation:0.11157
  |
Correlation:0.068967
  |
Correlation:0.38158
  |
Correlation:0.38158
  |
Correlation:0.11157
  |
Component:313; Jaccard:0.10324
 |
Component:199; Jaccard:0.1063
 |
Component:250; Jaccard:0.11317
 |
Component:250; Jaccard:0.11121
 |
Component:146; Jaccard:0.091202
 |
Component:146; Jaccard:0.061591
 |
Component:389; Jaccard:0.037131
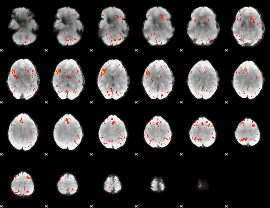 |
Correlation:0.11157
  |
Correlation:0.14377
  |
Correlation:0.38158
  |
Correlation:0.38158
  |
Correlation:0.068967
  |
Correlation:0.068967
  |
Correlation:0.1289
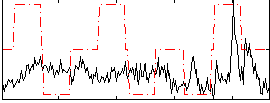 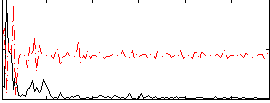 |
Component:203; Jaccard:0.094638
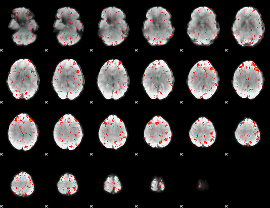 |
Component:250; Jaccard:0.10319
 |
Component:203; Jaccard:0.10743
 |
Component:203; Jaccard:0.1014
 |
Component:203; Jaccard:0.081315
 |
Component:389; Jaccard:0.061537
 |
Component:146; Jaccard:0.036731
 |
Correlation:0.22054
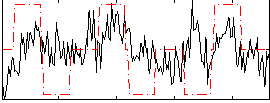 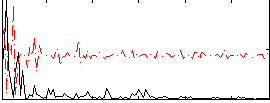 |
Correlation:0.38158
  |
Correlation:0.22054
  |
Correlation:0.22054
  |
Correlation:0.22054
  |
Correlation:0.1289
  |
Correlation:0.068967
  |
Component:250; Jaccard:0.090041
 |
Component:203; Jaccard:0.10292
 |
Component:199; Jaccard:0.098947
 |
Component:389; Jaccard:0.091013
 |
Component:389; Jaccard:0.078864
 |
Component:203; Jaccard:0.055063
 |
Component:385; Jaccard:0.035132
 |
Correlation:0.38158
  |
Correlation:0.22054
  |
Correlation:0.14377
  |
Correlation:0.1289
  |
Correlation:0.1289
  |
Correlation:0.22054
  |
Correlation:0.14845
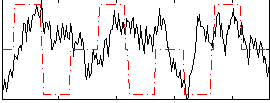 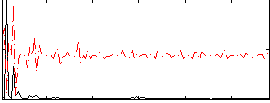 |
Component:385; Jaccard:0.089302
 |
Component:385; Jaccard:0.095367
 |
Component:303; Jaccard:0.096008
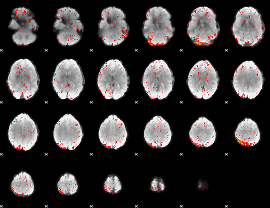 |
Component:303; Jaccard:0.08514
 |
Component:303; Jaccard:0.069247
 |
Component:385; Jaccard:0.052652
 |
Component:203; Jaccard:0.030516
 |
Correlation:0.14845
  |
Correlation:0.14845
  |
Correlation:0.10037
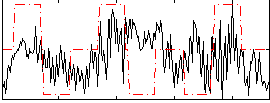 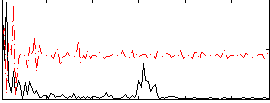 |
Correlation:0.10037
  |
Correlation:0.10037
  |
Correlation:0.14845
  |
Correlation:0.22054
  |
Cope: 05:"neg_MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
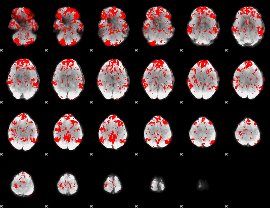 |
GLM binary result thresholded by 1.5
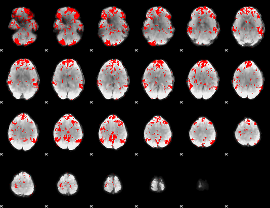 |
GLM binary result thresholded by 2
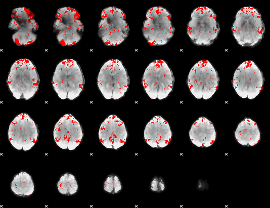 |
GLM binary result thresholded by 2.5
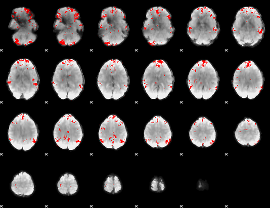 |
GLM binary result thresholded by 3
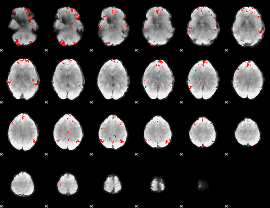 |
GLM binary result thresholded by 3.5
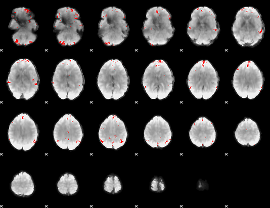 |
GLM binary result thresholded by 4
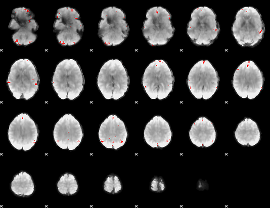 |
GLM Z-score result thresholded by 1
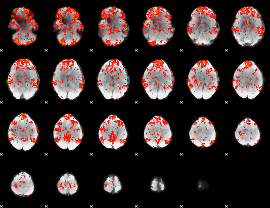 |
GLM Z-score result thresholded by 1.5
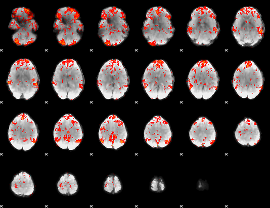 |
GLM Z-score result thresholded by 2
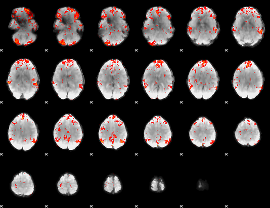 |
GLM Z-score result thresholded by 2.5
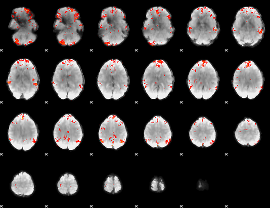 |
GLM Z-score result thresholded by 3
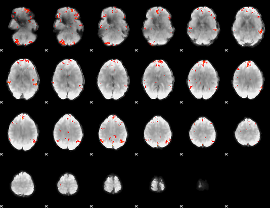 |
GLM Z-score result thresholded by 3.5
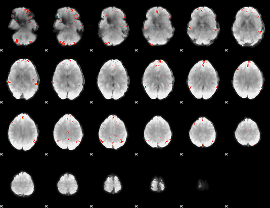 |
GLM Z-score result thresholded by 4
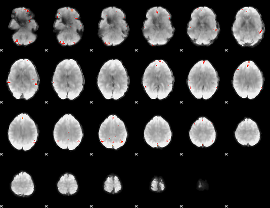 |
Component:050; Jaccard:0.11957
 |
Component:283; Jaccard:0.14063
 |
Component:283; Jaccard:0.1553
 |
Component:283; Jaccard:0.15027
 |
Component:394; Jaccard:0.12352
 |
Component:394; Jaccard:0.090663
 |
Component:394; Jaccard:0.051607
 |
Correlation:0.035571
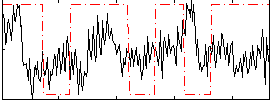 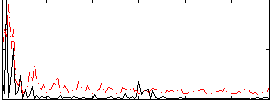 |
Correlation:0.11753
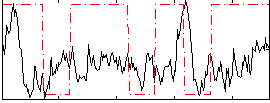 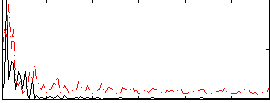 |
Correlation:0.11753
  |
Correlation:0.11753
  |
Correlation:-0.059288
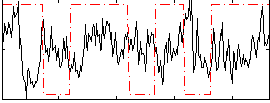 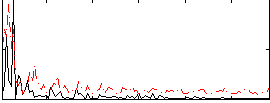 |
Correlation:-0.059288
  |
Correlation:-0.059288
  |
Component:394; Jaccard:0.1165
 |
Component:394; Jaccard:0.13741
 |
Component:394; Jaccard:0.15066
 |
Component:394; Jaccard:0.14546
 |
Component:283; Jaccard:0.12161
 |
Component:208; Jaccard:0.084342
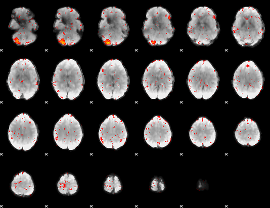 |
Component:208; Jaccard:0.051252
 |
Correlation:-0.059288
  |
Correlation:-0.059288
  |
Correlation:-0.059288
  |
Correlation:-0.059288
  |
Correlation:0.11753
  |
Correlation:0.31178
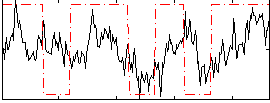  |
Correlation:0.31178
  |
Component:283; Jaccard:0.11618
 |
Component:050; Jaccard:0.13485
 |
Component:050; Jaccard:0.14274
 |
Component:208; Jaccard:0.13029
 |
Component:208; Jaccard:0.11689
 |
Component:283; Jaccard:0.081773
 |
Component:283; Jaccard:0.042897
 |
Correlation:0.11753
  |
Correlation:0.035571
  |
Correlation:0.035571
  |
Correlation:0.31178
  |
Correlation:0.31178
  |
Correlation:0.11753
  |
Correlation:0.11753
  |
Component:208; Jaccard:0.10348
 |
Component:208; Jaccard:0.11948
 |
Component:208; Jaccard:0.13185
 |
Component:050; Jaccard:0.12952
 |
Component:050; Jaccard:0.10092
 |
Component:050; Jaccard:0.066499
 |
Component:050; Jaccard:0.036185
 |
Correlation:0.31178
  |
Correlation:0.31178
  |
Correlation:0.31178
  |
Correlation:0.035571
  |
Correlation:0.035571
  |
Correlation:0.035571
  |
Correlation:0.035571
  |
Component:315; Jaccard:0.099103
 |
Component:319; Jaccard:0.11402
 |
Component:319; Jaccard:0.12185
 |
Component:319; Jaccard:0.11516
 |
Component:319; Jaccard:0.089279
 |
Component:319; Jaccard:0.059143
 |
Component:305; Jaccard:0.034438
 |
Correlation:0.050476
 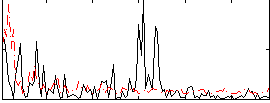 |
Correlation:-0.13168
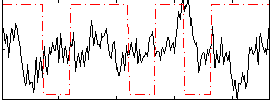 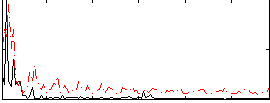 |
Correlation:-0.13168
  |
Correlation:-0.13168
  |
Correlation:-0.13168
  |
Correlation:-0.13168
  |
Correlation:0.061182
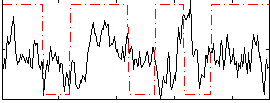 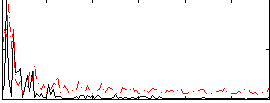 |
Component:305; Jaccard:0.098998
 |
Component:315; Jaccard:0.1111
 |
Component:305; Jaccard:0.11357
 |
Component:397; Jaccard:0.10643
 |
Component:397; Jaccard:0.083206
 |
Component:305; Jaccard:0.058267
 |
Component:397; Jaccard:0.031482
 |
Correlation:0.061182
  |
Correlation:0.050476
  |
Correlation:0.061182
  |
Correlation:0.1254
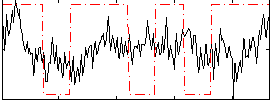 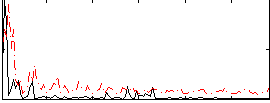 |
Correlation:0.1254
  |
Correlation:0.061182
  |
Correlation:0.1254
  |
Component:234; Jaccard:0.098229
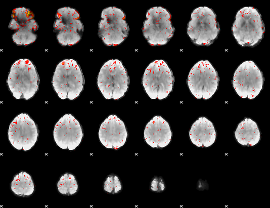 |
Component:305; Jaccard:0.11006
 |
Component:397; Jaccard:0.11157
 |
Component:305; Jaccard:0.10344
 |
Component:305; Jaccard:0.081377
 |
Component:397; Jaccard:0.057687
 |
Component:319; Jaccard:0.027736
 |
Correlation:0.093897
  |
Correlation:0.061182
  |
Correlation:0.1254
  |
Correlation:0.061182
  |
Correlation:0.061182
  |
Correlation:0.1254
  |
Correlation:-0.13168
  |
Component:311; Jaccard:0.097949
 |
Component:311; Jaccard:0.10776
 |
Component:315; Jaccard:0.11049
 |
Component:311; Jaccard:0.10058
 |
Component:311; Jaccard:0.078244
 |
Component:311; Jaccard:0.05115
 |
Component:311; Jaccard:0.027023
 |
Correlation:-0.066488
  |
Correlation:-0.066488
  |
Correlation:0.050476
  |
Correlation:-0.066488
  |
Correlation:-0.066488
  |
Correlation:-0.066488
  |
Correlation:-0.066488
  |
Component:319; Jaccard:0.097721
 |
Component:397; Jaccard:0.10583
 |
Component:311; Jaccard:0.11
 |
Component:315; Jaccard:0.098869
 |
Component:315; Jaccard:0.072729
 |
Component:315; Jaccard:0.049194
 |
Component:337; Jaccard:0.024617
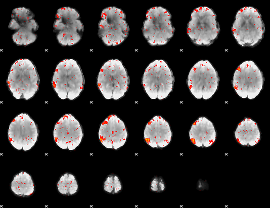 |
Correlation:-0.13168
  |
Correlation:0.1254
  |
Correlation:-0.066488
  |
Correlation:0.050476
  |
Correlation:0.050476
  |
Correlation:0.050476
  |
Correlation:0.035056
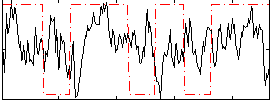 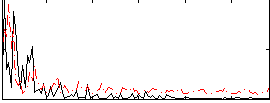 |
Component:265; Jaccard:0.096761
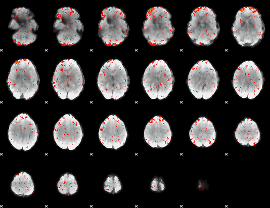 |
Component:234; Jaccard:0.10487
 |
Component:265; Jaccard:0.10395
 |
Component:256; Jaccard:0.093186
 |
Component:256; Jaccard:0.069527
 |
Component:256; Jaccard:0.045333
 |
Component:322; Jaccard:0.022908
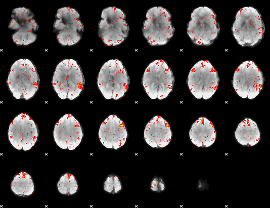 |
Correlation:0.1686
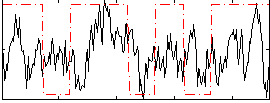 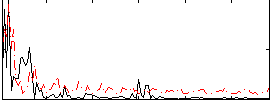 |
Correlation:0.093897
  |
Correlation:0.1686
  |
Correlation:-0.0015865
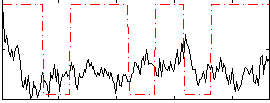  |
Correlation:-0.0015865
  |
Correlation:-0.0015865
  |
Correlation:-0.042345
  |
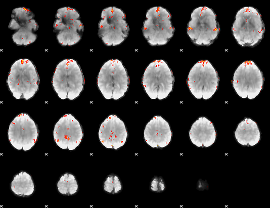
Cope: 06:"neg_REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
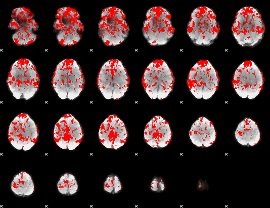 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
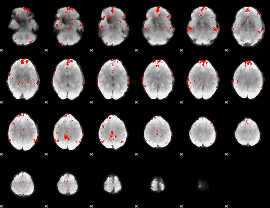 |
GLM binary result thresholded by 4
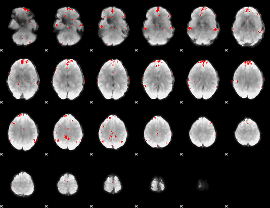 |
GLM Z-score result thresholded by 1
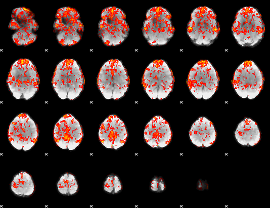 |
GLM Z-score result thresholded by 1.5
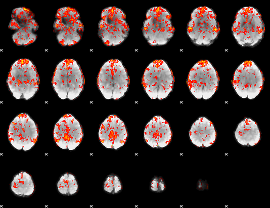 |
GLM Z-score result thresholded by 2
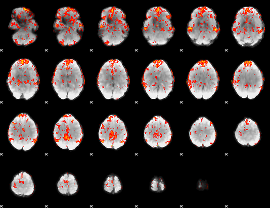 |
GLM Z-score result thresholded by 2.5
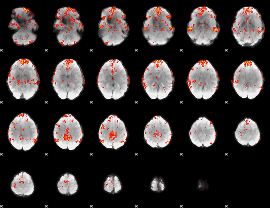 |
GLM Z-score result thresholded by 3
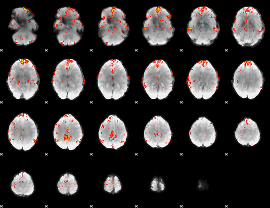 |
GLM Z-score result thresholded by 3.5
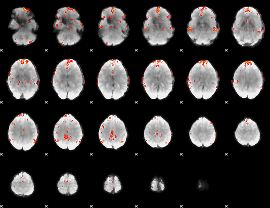 |
GLM Z-score result thresholded by 4
 |
Component:311; Jaccard:0.1049
 |
Component:311; Jaccard:0.12071
 |
Component:311; Jaccard:0.13665
 |
Component:256; Jaccard:0.15385
 |
Component:319; Jaccard:0.16846
 |
Component:256; Jaccard:0.16701
 |
Component:256; Jaccard:0.15366
 |
Correlation:0.25145
  |
Correlation:0.25145
  |
Correlation:0.25145
  |
Correlation:0.32402
 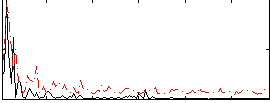 |
Correlation:0.37076
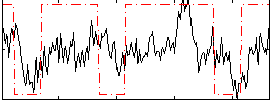 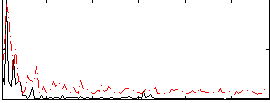 |
Correlation:0.32402
  |
Correlation:0.32402
  |
Component:308; Jaccard:0.10428
 |
Component:050; Jaccard:0.11506
 |
Component:319; Jaccard:0.13411
 |
Component:319; Jaccard:0.15266
 |
Component:256; Jaccard:0.16738
 |
Component:319; Jaccard:0.16466
 |
Component:319; Jaccard:0.14657
 |
Correlation:0.26805
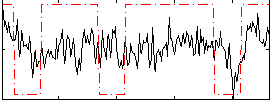  |
Correlation:-0.020316
  |
Correlation:0.37076
  |
Correlation:0.37076
  |
Correlation:0.32402
  |
Correlation:0.37076
  |
Correlation:0.37076
  |
Component:050; Jaccard:0.10269
 |
Component:308; Jaccard:0.11375
 |
Component:256; Jaccard:0.13336
 |
Component:311; Jaccard:0.14872
 |
Component:311; Jaccard:0.15298
 |
Component:311; Jaccard:0.14629
 |
Component:311; Jaccard:0.12918
 |
Correlation:-0.020316
  |
Correlation:0.26805
  |
Correlation:0.32402
  |
Correlation:0.25145
  |
Correlation:0.25145
  |
Correlation:0.25145
  |
Correlation:0.25145
  |
Component:283; Jaccard:0.096913
 |
Component:256; Jaccard:0.11276
 |
Component:283; Jaccard:0.12754
 |
Component:283; Jaccard:0.13986
 |
Component:283; Jaccard:0.14689
 |
Component:283; Jaccard:0.13709
 |
Component:283; Jaccard:0.11913
 |
Correlation:0.15122
  |
Correlation:0.32402
  |
Correlation:0.15122
  |
Correlation:0.15122
  |
Correlation:0.15122
  |
Correlation:0.15122
  |
Correlation:0.15122
  |
Component:256; Jaccard:0.093912
 |
Component:319; Jaccard:0.11254
 |
Component:050; Jaccard:0.12531
 |
Component:050; Jaccard:0.13316
 |
Component:050; Jaccard:0.13194
 |
Component:050; Jaccard:0.12272
 |
Component:050; Jaccard:0.10624
 |
Correlation:0.32402
  |
Correlation:0.37076
  |
Correlation:-0.020316
  |
Correlation:-0.020316
  |
Correlation:-0.020316
  |
Correlation:-0.020316
  |
Correlation:-0.020316
  |
Component:319; Jaccard:0.09386
 |
Component:283; Jaccard:0.11032
 |
Component:308; Jaccard:0.12009
 |
Component:308; Jaccard:0.12355
 |
Component:075; Jaccard:0.12171
 |
Component:075; Jaccard:0.11307
 |
Component:075; Jaccard:0.099219
 |
Correlation:0.37076
  |
Correlation:0.15122
  |
Correlation:0.26805
  |
Correlation:0.26805
  |
Correlation:-0.021318
  |
Correlation:-0.021318
  |
Correlation:-0.021318
  |
Component:281; Jaccard:0.093559
 |
Component:075; Jaccard:0.10319
 |
Component:075; Jaccard:0.11203
 |
Component:075; Jaccard:0.11924
 |
Component:308; Jaccard:0.11718
 |
Component:397; Jaccard:0.10787
 |
Component:397; Jaccard:0.094732
 |
Correlation:0.37965
  |
Correlation:-0.021318
  |
Correlation:-0.021318
  |
Correlation:-0.021318
  |
Correlation:0.26805
  |
Correlation:-0.082514
  |
Correlation:-0.082514
  |
Component:075; Jaccard:0.09249
 |
Component:251; Jaccard:0.101
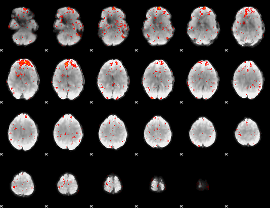 |
Component:397; Jaccard:0.11195
 |
Component:397; Jaccard:0.11654
 |
Component:397; Jaccard:0.11689
 |
Component:308; Jaccard:0.10171
 |
Component:394; Jaccard:0.082137
 |
Correlation:-0.021318
  |
Correlation:0.038813
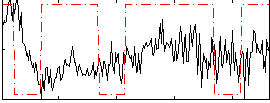 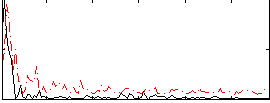 |
Correlation:-0.082514
  |
Correlation:-0.082514
  |
Correlation:-0.082514
  |
Correlation:0.26805
  |
Correlation:0.15599
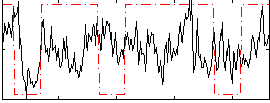  |
Component:251; Jaccard:0.091895
 |
Component:281; Jaccard:0.10085
 |
Component:262; Jaccard:0.10964
 |
Component:394; Jaccard:0.11517
 |
Component:251; Jaccard:0.11296
 |
Component:251; Jaccard:0.10103
 |
Component:251; Jaccard:0.079816
 |
Correlation:0.038813
  |
Correlation:0.37965
  |
Correlation:0.13011
  |
Correlation:0.15599
  |
Correlation:0.038813
  |
Correlation:0.038813
  |
Correlation:0.038813
  |
Component:394; Jaccard:0.089368
 |
Component:397; Jaccard:0.099534
 |
Component:394; Jaccard:0.10928
 |
Component:251; Jaccard:0.11374
 |
Component:394; Jaccard:0.11043
 |
Component:394; Jaccard:0.10042
 |
Component:308; Jaccard:0.07676
 |
Correlation:0.15599
  |
Correlation:-0.082514
  |
Correlation:0.15599
  |
Correlation:0.038813
  |
Correlation:0.15599
  |
Correlation:0.15599
  |
Correlation:0.26805
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































