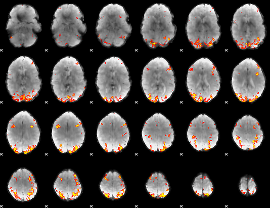
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
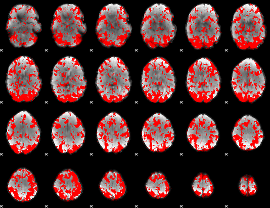 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
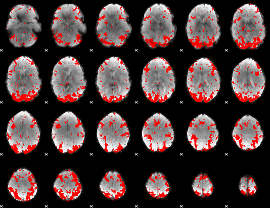 |
GLM binary result thresholded by 2.5
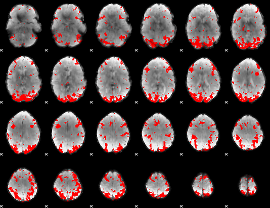 |
GLM binary result thresholded by 3
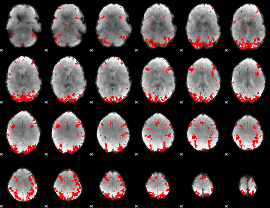 |
GLM binary result thresholded by 3.5
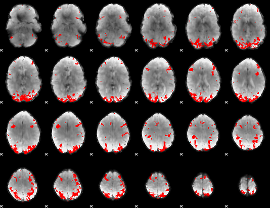 |
GLM binary result thresholded by 4
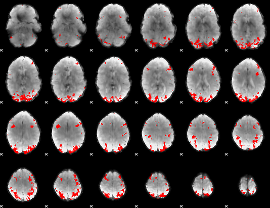 |
GLM Z-score result thresholded by 1
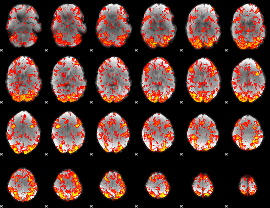 |
GLM Z-score result thresholded by 1.5
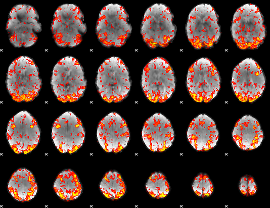 |
GLM Z-score result thresholded by 2
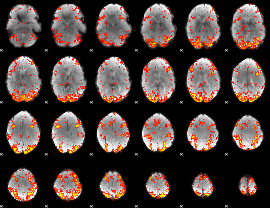 |
GLM Z-score result thresholded by 2.5
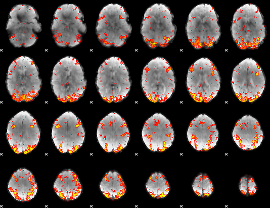 |
GLM Z-score result thresholded by 3
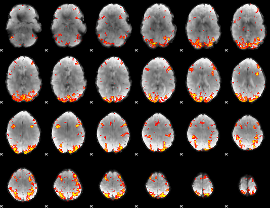 |
GLM Z-score result thresholded by 3.5
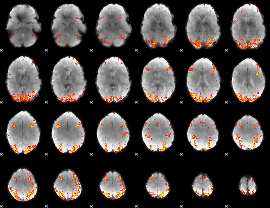 |
GLM Z-score result thresholded by 4
 |
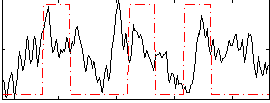
Component:071; Jaccard:0.13412
 |
Component:181; Jaccard:0.16939
 |
Component:181; Jaccard:0.21685
 |
Component:181; Jaccard:0.26343
 |
Component:181; Jaccard:0.29987
 |
Component:181; Jaccard:0.31739
 |
Component:181; Jaccard:0.31646
 |
Correlation:0.22682
  |
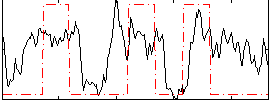
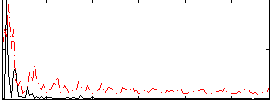
Correlation:0.34526
  |
Correlation:0.34526
  |
Correlation:0.34526
  |
Correlation:0.34526
  |
Correlation:0.34526
  |
Correlation:0.34526
  |
Component:181; Jaccard:0.13016
 |
Component:071; Jaccard:0.1672
 |
Component:071; Jaccard:0.20242
 |
Component:071; Jaccard:0.23461
 |
Component:071; Jaccard:0.25493
 |
Component:071; Jaccard:0.26264
 |
Component:071; Jaccard:0.25573
 |
Correlation:0.34526
  |
Correlation:0.22682
  |
Correlation:0.22682
  |
Correlation:0.22682
  |
Correlation:0.22682
  |
Correlation:0.22682
  |
Correlation:0.22682
  |
Component:292; Jaccard:0.12096
 |
Component:292; Jaccard:0.15177
 |
Component:292; Jaccard:0.1833
 |
Component:292; Jaccard:0.21009
 |
Component:292; Jaccard:0.22875
 |
Component:292; Jaccard:0.23228
 |
Component:292; Jaccard:0.22165
 |
Correlation:0.26105
  |
Correlation:0.26105
  |
Correlation:0.26105
  |
Correlation:0.26105
  |
Correlation:0.26105
  |
Correlation:0.26105
  |
Correlation:0.26105
  |
Component:268; Jaccard:0.10846
 |
Component:335; Jaccard:0.1342
 |
Component:335; Jaccard:0.16471
 |
Component:335; Jaccard:0.19275
 |
Component:335; Jaccard:0.21523
 |
Component:335; Jaccard:0.2234
 |
Component:335; Jaccard:0.21834
 |
Correlation:0.16503
  |
Correlation:0.2219
  |
Correlation:0.2219
  |
Correlation:0.2219
  |
Correlation:0.2219
  |
Correlation:0.2219
  |
Correlation:0.2219
  |
Component:309; Jaccard:0.10737
 |
Component:182; Jaccard:0.12604
 |
Component:182; Jaccard:0.14769
 |
Component:182; Jaccard:0.16534
 |
Component:182; Jaccard:0.17535
 |
Component:182; Jaccard:0.17355
 |
Component:182; Jaccard:0.16483
 |
Correlation:0.15283
  |
Correlation:0.18671
  |
Correlation:0.18671
  |
Correlation:0.18671
  |
Correlation:0.18671
  |
Correlation:0.18671
  |
Correlation:0.18671
  |
Component:335; Jaccard:0.10728
 |
Component:309; Jaccard:0.1257
 |
Component:075; Jaccard:0.14408
 |
Component:075; Jaccard:0.16114
 |
Component:075; Jaccard:0.17058
 |
Component:075; Jaccard:0.17172
 |
Component:075; Jaccard:0.16178
 |
Correlation:0.2219
  |
Correlation:0.15283
  |
Correlation:0.039592
  |
Correlation:0.039592
  |
Correlation:0.039592
  |
Correlation:0.039592
  |
Correlation:0.039592
  |
Component:070; Jaccard:0.10612
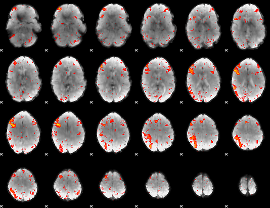 |
Component:075; Jaccard:0.12443
 |
Component:309; Jaccard:0.14338
 |
Component:309; Jaccard:0.15803
 |
Component:309; Jaccard:0.16383
 |
Component:309; Jaccard:0.1596
 |
Component:150; Jaccard:0.15105
 |
Correlation:0.095954
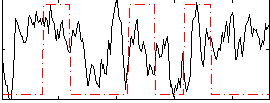 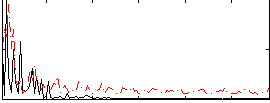 |
Correlation:0.039592
  |
Correlation:0.15283
  |
Correlation:0.15283
  |
Correlation:0.15283
  |
Correlation:0.15283
  |
Correlation:0.32756
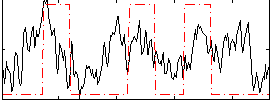  |
Component:091; Jaccard:0.10458
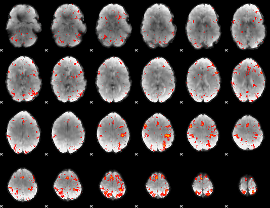 |
Component:268; Jaccard:0.12381
 |
Component:091; Jaccard:0.13502
 |
Component:150; Jaccard:0.14556
 |
Component:150; Jaccard:0.15573
 |
Component:150; Jaccard:0.15896
 |
Component:308; Jaccard:0.15001
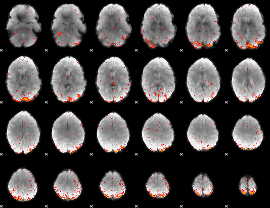 |
Correlation:0.14833
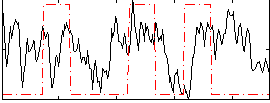 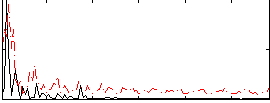 |
Correlation:0.16503
  |
Correlation:0.14833
  |
Correlation:0.32756
  |
Correlation:0.32756
  |
Correlation:0.32756
  |
Correlation:0.58261
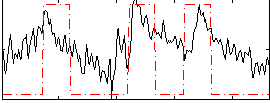  |
Component:182; Jaccard:0.10452
 |
Component:070; Jaccard:0.12195
 |
Component:070; Jaccard:0.1348
 |
Component:091; Jaccard:0.14317
 |
Component:308; Jaccard:0.15178
 |
Component:308; Jaccard:0.15527
 |
Component:309; Jaccard:0.14715
 |
Correlation:0.18671
  |
Correlation:0.095954
  |
Correlation:0.095954
  |
Correlation:0.14833
  |
Correlation:0.58261
  |
Correlation:0.58261
  |
Correlation:0.15283
  |
Component:075; Jaccard:0.10437
 |
Component:091; Jaccard:0.12109
 |
Component:268; Jaccard:0.13245
 |
Component:308; Jaccard:0.14164
 |
Component:091; Jaccard:0.14486
 |
Component:091; Jaccard:0.13764
 |
Component:091; Jaccard:0.12692
 |
Correlation:0.039592
  |
Correlation:0.14833
  |
Correlation:0.16503
  |
Correlation:0.58261
  |
Correlation:0.14833
  |
Correlation:0.14833
  |
Correlation:0.14833
  |
Cope: 02:"REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
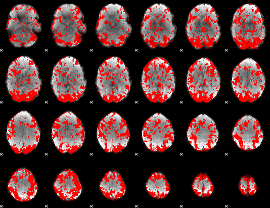 |
GLM binary result thresholded by 1.5
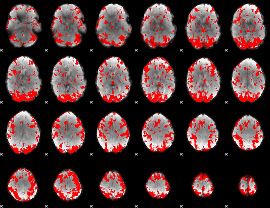 |
GLM binary result thresholded by 2
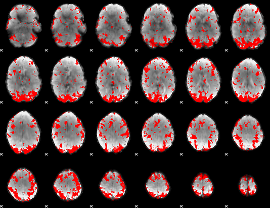 |
GLM binary result thresholded by 2.5
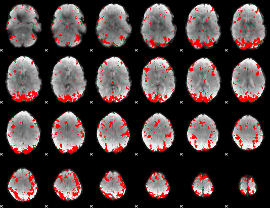 |
GLM binary result thresholded by 3
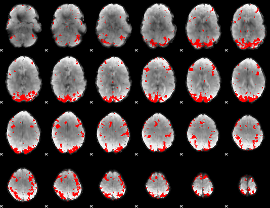 |
GLM binary result thresholded by 3.5
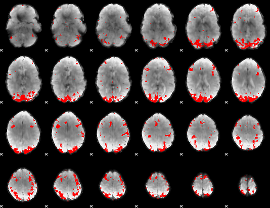 |
GLM binary result thresholded by 4
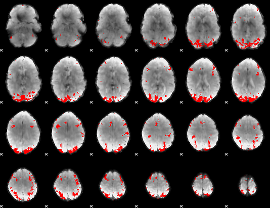 |
GLM Z-score result thresholded by 1
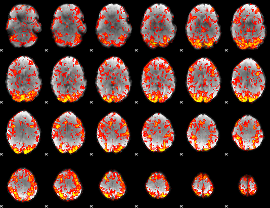 |
GLM Z-score result thresholded by 1.5
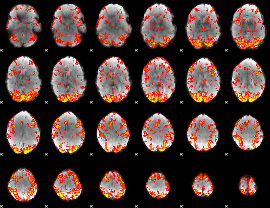 |
GLM Z-score result thresholded by 2
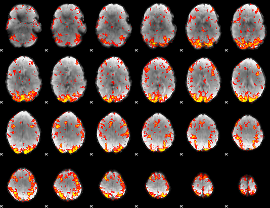 |
GLM Z-score result thresholded by 2.5
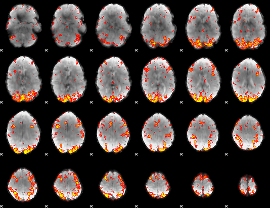 |
GLM Z-score result thresholded by 3
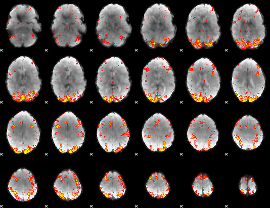 |
GLM Z-score result thresholded by 3.5
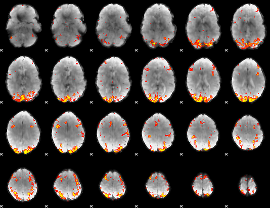 |
GLM Z-score result thresholded by 4
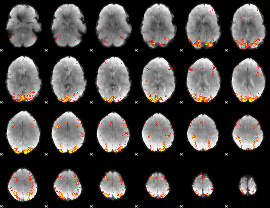 |
Component:071; Jaccard:0.13236
 |
Component:071; Jaccard:0.16424
 |
Component:071; Jaccard:0.19854
 |
Component:071; Jaccard:0.23163
 |
Component:071; Jaccard:0.25632
 |
Component:071; Jaccard:0.26569
 |
Component:071; Jaccard:0.26109
 |
Correlation:0.067177
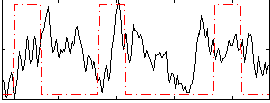  |
Correlation:0.067177
  |
Correlation:0.067177
  |
Correlation:0.067177
  |
Correlation:0.067177
  |
Correlation:0.067177
  |
Correlation:0.067177
  |
Component:075; Jaccard:0.12717
 |
Component:075; Jaccard:0.1588
 |
Component:075; Jaccard:0.19273
 |
Component:075; Jaccard:0.22301
 |
Component:075; Jaccard:0.24228
 |
Component:075; Jaccard:0.25314
 |
Component:075; Jaccard:0.24808
 |
Correlation:0.34359
  |
Correlation:0.34359
  |
Correlation:0.34359
  |
Correlation:0.34359
  |
Correlation:0.34359
  |
Correlation:0.34359
  |
Correlation:0.34359
  |
Component:181; Jaccard:0.12154
 |
Component:181; Jaccard:0.15242
 |
Component:181; Jaccard:0.18783
 |
Component:181; Jaccard:0.22165
 |
Component:181; Jaccard:0.24151
 |
Component:181; Jaccard:0.24843
 |
Component:181; Jaccard:0.23773
 |
Correlation:0.12037
 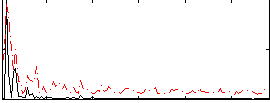 |
Correlation:0.12037
  |
Correlation:0.12037
  |
Correlation:0.12037
  |
Correlation:0.12037
  |
Correlation:0.12037
  |
Correlation:0.12037
  |
Component:292; Jaccard:0.11774
 |
Component:292; Jaccard:0.14609
 |
Component:292; Jaccard:0.17632
 |
Component:292; Jaccard:0.20205
 |
Component:292; Jaccard:0.21982
 |
Component:292; Jaccard:0.22289
 |
Component:292; Jaccard:0.21479
 |
Correlation:0.22283
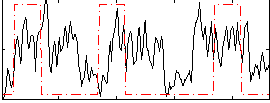 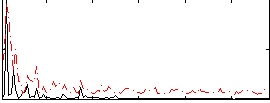 |
Correlation:0.22283
  |
Correlation:0.22283
  |
Correlation:0.22283
  |
Correlation:0.22283
  |
Correlation:0.22283
  |
Correlation:0.22283
  |
Component:182; Jaccard:0.11402
 |
Component:182; Jaccard:0.14114
 |
Component:182; Jaccard:0.16746
 |
Component:182; Jaccard:0.19034
 |
Component:182; Jaccard:0.19989
 |
Component:335; Jaccard:0.20579
 |
Component:335; Jaccard:0.20127
 |
Correlation:0.23957
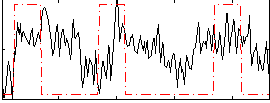  |
Correlation:0.23957
  |
Correlation:0.23957
  |
Correlation:0.23957
  |
Correlation:0.23957
  |
Correlation:0.091731
  |
Correlation:0.091731
  |
Component:309; Jaccard:0.10731
 |
Component:335; Jaccard:0.12701
 |
Component:335; Jaccard:0.15328
 |
Component:335; Jaccard:0.17986
 |
Component:335; Jaccard:0.19835
 |
Component:182; Jaccard:0.19849
 |
Component:150; Jaccard:0.1956
 |
Correlation:0.085539
  |
Correlation:0.091731
  |
Correlation:0.091731
  |
Correlation:0.091731
  |
Correlation:0.091731
  |
Correlation:0.23957
  |
Correlation:0.24557
  |
Component:344; Jaccard:0.10584
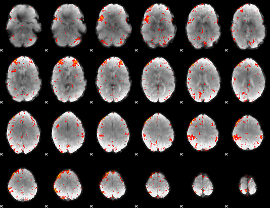 |
Component:309; Jaccard:0.12451
 |
Component:150; Jaccard:0.14778
 |
Component:150; Jaccard:0.17148
 |
Component:150; Jaccard:0.1889
 |
Component:150; Jaccard:0.19795
 |
Component:182; Jaccard:0.19071
 |
Correlation:0.13799
  |
Correlation:0.085539
  |
Correlation:0.24557
  |
Correlation:0.24557
  |
Correlation:0.24557
  |
Correlation:0.24557
  |
Correlation:0.23957
  |
Component:268; Jaccard:0.1052
 |
Component:150; Jaccard:0.12272
 |
Component:309; Jaccard:0.14087
 |
Component:309; Jaccard:0.15291
 |
Component:309; Jaccard:0.15702
 |
Component:309; Jaccard:0.15089
 |
Component:309; Jaccard:0.13846
 |
Correlation:-0.049645
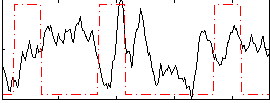  |
Correlation:0.24557
  |
Correlation:0.085539
  |
Correlation:0.085539
  |
Correlation:0.085539
  |
Correlation:0.085539
  |
Correlation:0.085539
  |
Component:335; Jaccard:0.10326
 |
Component:268; Jaccard:0.11887
 |
Component:268; Jaccard:0.1277
 |
Component:070; Jaccard:0.13082
 |
Component:308; Jaccard:0.13141
 |
Component:308; Jaccard:0.13204
 |
Component:308; Jaccard:0.12637
 |
Correlation:0.091731
  |
Correlation:-0.049645
  |
Correlation:-0.049645
  |
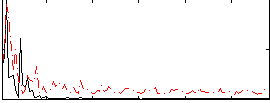
Correlation:0.2503
 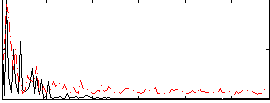 |
Correlation:-0.21916
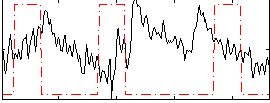  |
Correlation:-0.21916
  |
Correlation:-0.21916
  |
Component:070; Jaccard:0.10104
 |
Component:344; Jaccard:0.11826
 |
Component:070; Jaccard:0.12577
 |
Component:091; Jaccard:0.13065
 |
Component:070; Jaccard:0.12839
 |
Component:091; Jaccard:0.12075
 |
Component:091; Jaccard:0.11097
 |
Correlation:0.2503
  |
Correlation:0.13799
  |
Correlation:0.2503
  |
Correlation:0.11967
  |
Correlation:0.2503
  |
Correlation:0.11967
  |
Correlation:0.11967
  |
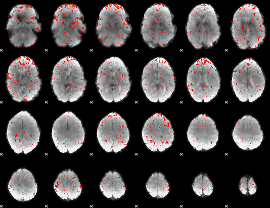
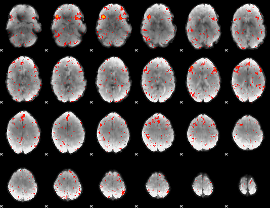
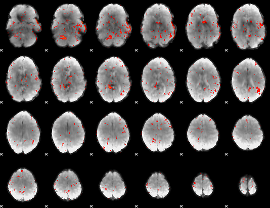
Cope: 03:"MATCH-REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
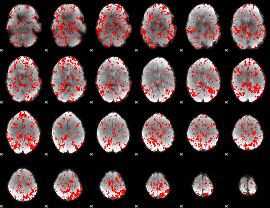 |
GLM binary result thresholded by 1.5
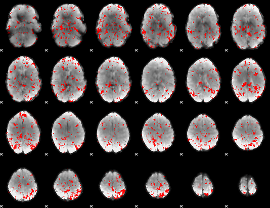 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
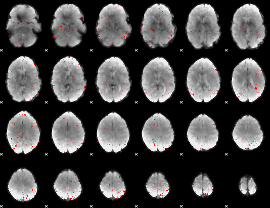 |
GLM binary result thresholded by 3
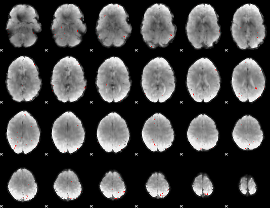 |
GLM binary result thresholded by 3.5
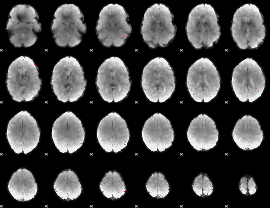 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
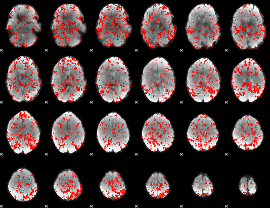 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
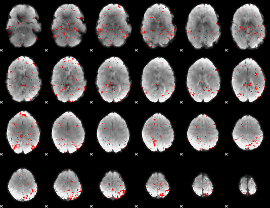 |
GLM Z-score result thresholded by 2.5
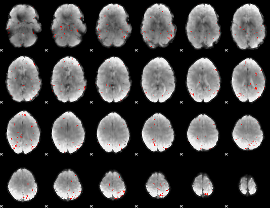 |
GLM Z-score result thresholded by 3
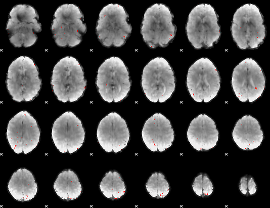 |
GLM Z-score result thresholded by 3.5
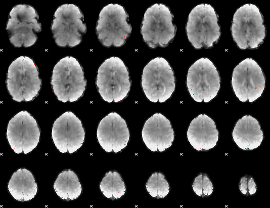 |
GLM Z-score result thresholded by 4
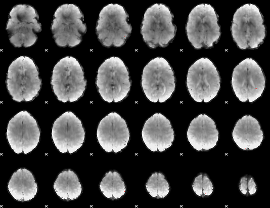 |
Component:341; Jaccard:0.092682
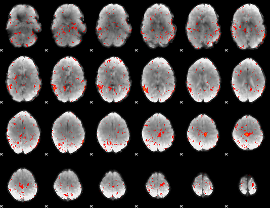 |
Component:341; Jaccard:0.095502
 |
Component:341; Jaccard:0.072312
 |
Component:039; Jaccard:0.036771
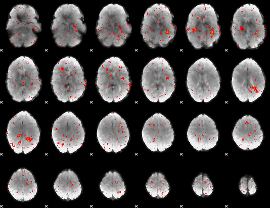 |
Component:039; Jaccard:0.011986
 |
Component:039; Jaccard:0.004929
 |
Component:039; Jaccard:0.001853
 |
Correlation:0.28886
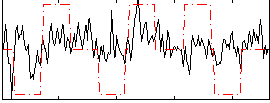 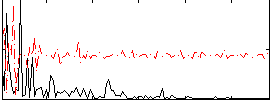 |
Correlation:0.28886
  |
Correlation:0.28886
  |
Correlation:0.20107
 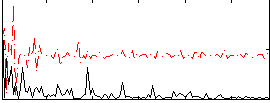 |
Correlation:0.20107
  |
Correlation:0.20107
  |
Correlation:0.20107
  |
Component:022; Jaccard:0.084328
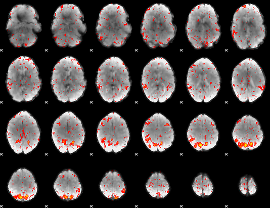 |
Component:283; Jaccard:0.081382
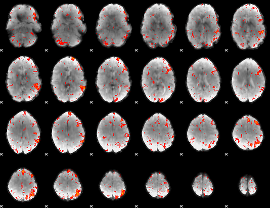 |
Component:199; Jaccard:0.063779
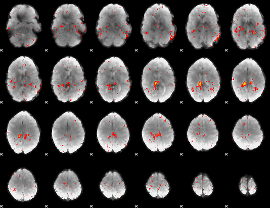 |
Component:341; Jaccard:0.035974
 |
Component:181; Jaccard:0.011151
 |
Component:199; Jaccard:0.00374
 |
Component:275; Jaccard:0.001487
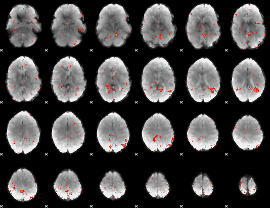 |
Correlation:0.32808
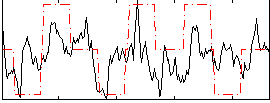 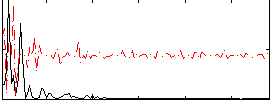 |
Correlation:0.13101
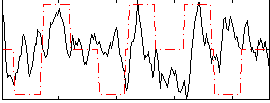 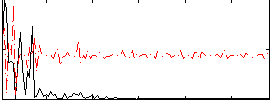 |
Correlation:0.26832
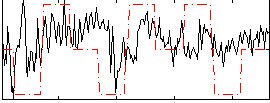  |
Correlation:0.28886
  |
Correlation:0.1333
  |
Correlation:0.26832
  |
Correlation:0.17893
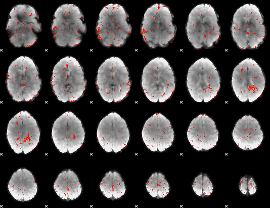
 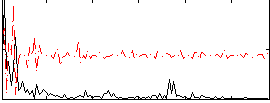 |
Component:283; Jaccard:0.083059
 |
Component:324; Jaccard:0.076929
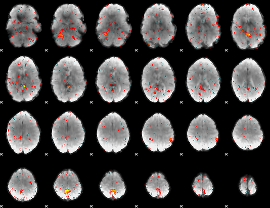
 |
Component:039; Jaccard:0.063494
 |
Component:181; Jaccard:0.035095
 |
Component:199; Jaccard:0.010983
 |
Component:036; Jaccard:0.003163
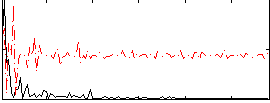
 |
Component:199; Jaccard:0.001423
 |
Correlation:0.13101
  |
Correlation:0.32941
  |
Correlation:0.20107
  |
Correlation:0.1333
  |
Correlation:0.26832
  |
Correlation:0.28501
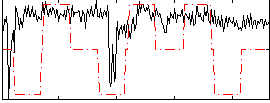 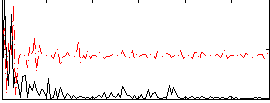 |
Correlation:0.26832
  |
Component:365; Jaccard:0.075595
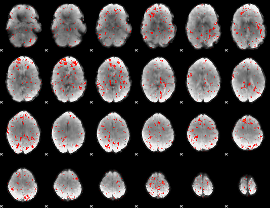 |
Component:022; Jaccard:0.076407
 |
Component:324; Jaccard:0.062893
 |
Component:199; Jaccard:0.033401
 |
Component:324; Jaccard:0.010586
 |
Component:324; Jaccard:0.003085
 |
Component:016; Jaccard:0.001343
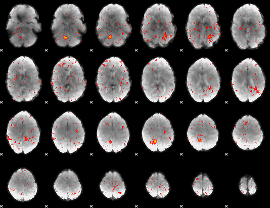 |
Correlation:0.10411
  |
Correlation:0.32808
  |
Correlation:0.32941
  |
Correlation:0.26832
  |
Correlation:0.32941
  |
Correlation:0.32941
  |
Correlation:0.25248
  |
Component:161; Jaccard:0.072622
 |
Component:181; Jaccard:0.074241
 |
Component:181; Jaccard:0.062504
 |
Component:324; Jaccard:0.032829
 |
Component:341; Jaccard:0.010213
 |
Component:380; Jaccard:0.003066
 |
Component:212; Jaccard:0.001144
 |
Correlation:0.22998
  |
Correlation:0.1333
  |
Correlation:0.1333
  |
Correlation:0.32941
  |
Correlation:0.28886
  |
Correlation:0.23341
 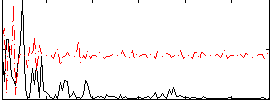 |
Correlation:0.010587
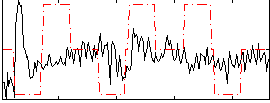 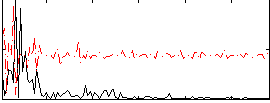 |
Component:380; Jaccard:0.072523
 |
Component:039; Jaccard:0.073518
 |
Component:283; Jaccard:0.059535
 |
Component:119; Jaccard:0.029243
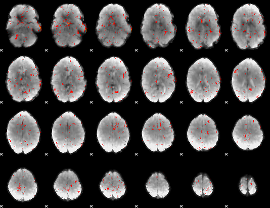 |
Component:016; Jaccard:0.009707
 |
Component:275; Jaccard:0.002912
 |
Component:036; Jaccard:0.001107
 |
Correlation:0.23341
  |
Correlation:0.20107
  |
Correlation:0.13101
  |
Correlation:0.19376
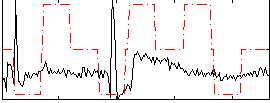 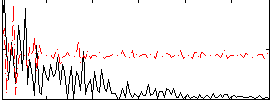 |
Correlation:0.25248
  |
Correlation:0.17893
  |
Correlation:0.28501
  |
Component:324; Jaccard:0.071811
 |
Component:380; Jaccard:0.071081
 |
Component:380; Jaccard:0.05414
 |
Component:283; Jaccard:0.02888
 |
Component:051; Jaccard:0.009655
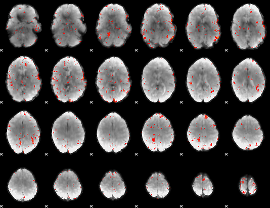 |
Component:341; Jaccard:0.002888
 |
Component:341; Jaccard:0.001068
 |
Correlation:0.32941
  |
Correlation:0.23341
  |
Correlation:0.23341
  |
Correlation:0.13101
  |
Correlation:0.30788
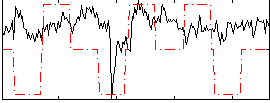  |
Correlation:0.28886
  |
Correlation:0.28886
  |
Component:181; Jaccard:0.069729
 |
Component:199; Jaccard:0.070945
 |
Component:022; Jaccard:0.052658
 |
Component:380; Jaccard:0.028026
 |
Component:380; Jaccard:0.009528
 |
Component:016; Jaccard:0.002795
 |
Component:051; Jaccard:0.001022
 |
Correlation:0.1333
  |
Correlation:0.26832
  |
Correlation:0.32808
  |
Correlation:0.23341
  |
Correlation:0.23341
  |
Correlation:0.25248
  |
Correlation:0.30788
  |
Component:172; Jaccard:0.069236
 |
Component:365; Jaccard:0.068824
 |
Component:172; Jaccard:0.052325
 |
Component:172; Jaccard:0.027738
 |
Component:129; Jaccard:0.009521
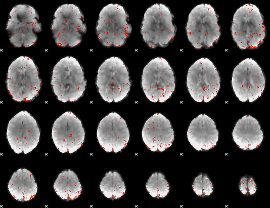 |
Component:272; Jaccard:0.002707
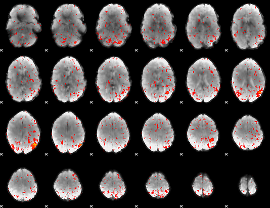 |
Component:324; Jaccard:0.000874
 |
Correlation:0.047568
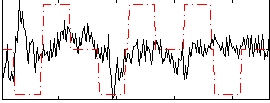 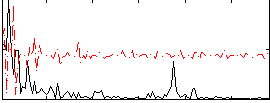 |
Correlation:0.10411
  |
Correlation:0.047568
  |
Correlation:0.047568
  |
Correlation:0.44038
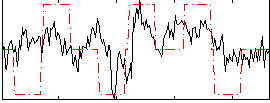 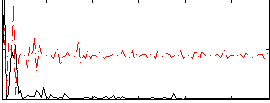 |
Correlation:0.14193
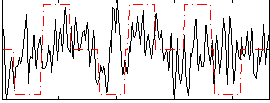 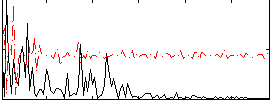 |
Correlation:0.32941
  |
Component:039; Jaccard:0.066904
 |
Component:172; Jaccard:0.068704
 |
Component:119; Jaccard:0.050542
 |
Component:022; Jaccard:0.025729
 |
Component:283; Jaccard:0.009425
 |
Component:181; Jaccard:0.002582
 |
Component:366; Jaccard:0.000843
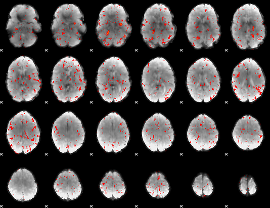 |
Correlation:0.20107
  |
Correlation:0.047568
  |
Correlation:0.19376
  |
Correlation:0.32808
  |
Correlation:0.13101
  |
Correlation:0.1333
  |
Correlation:0.058639
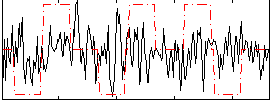  |
Cope: 04:"REL-MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
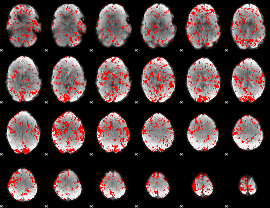 |
GLM binary result thresholded by 1.5
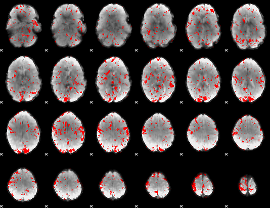 |
GLM binary result thresholded by 2
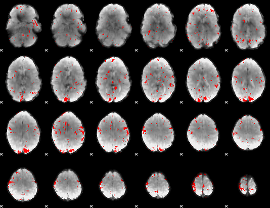 |
GLM binary result thresholded by 2.5
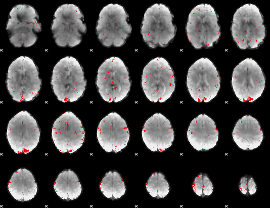 |
GLM binary result thresholded by 3
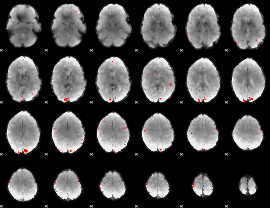 |
GLM binary result thresholded by 3.5
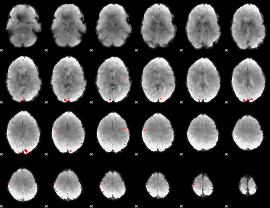 |
GLM binary result thresholded by 4
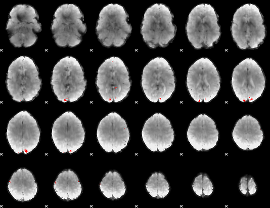 |
GLM Z-score result thresholded by 1
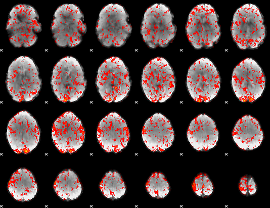 |
GLM Z-score result thresholded by 1.5
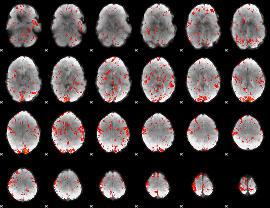 |
GLM Z-score result thresholded by 2
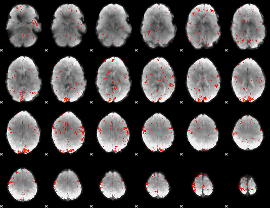 |
GLM Z-score result thresholded by 2.5
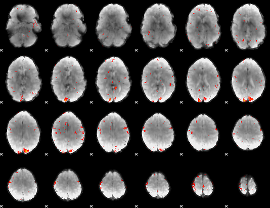 |
GLM Z-score result thresholded by 3
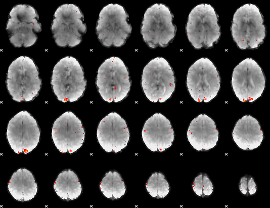 |
GLM Z-score result thresholded by 3.5
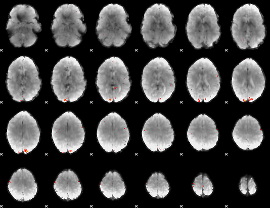 |
GLM Z-score result thresholded by 4
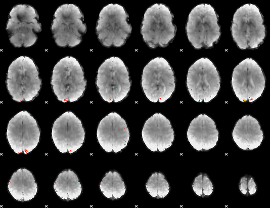 |
Component:075; Jaccard:0.12088
 |
Component:075; Jaccard:0.13452
 |
Component:075; Jaccard:0.12111
 |
Component:075; Jaccard:0.085498
 |
Component:075; Jaccard:0.052967
 |
Component:075; Jaccard:0.031606
 |
Component:075; Jaccard:0.020761
 |
Correlation:0.18018
  |
Correlation:0.18018
  |
Correlation:0.18018
  |
Correlation:0.18018
  |
Correlation:0.18018
  |
Correlation:0.18018
  |
Correlation:0.18018
  |
Component:391; Jaccard:0.097237
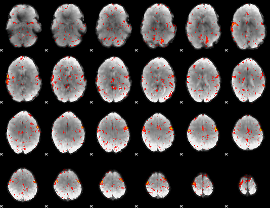 |
Component:391; Jaccard:0.097624
 |
Component:391; Jaccard:0.07706
 |
Component:391; Jaccard:0.048198
 |
Component:150; Jaccard:0.031126
 |
Component:150; Jaccard:0.019791
 |
Component:150; Jaccard:0.013451
 |
Correlation:0.3648
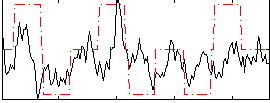 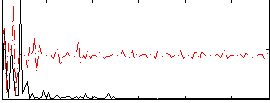 |
Correlation:0.3648
  |
Correlation:0.3648
  |
Correlation:0.3648
  |
Correlation:-0.048596
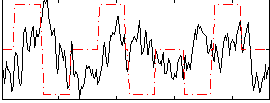 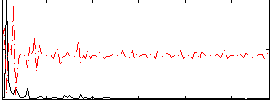 |
Correlation:-0.048596
  |
Correlation:-0.048596
  |
Component:337; Jaccard:0.075965
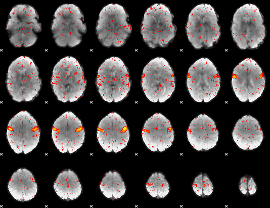 |
Component:337; Jaccard:0.075627
 |
Component:347; Jaccard:0.066143
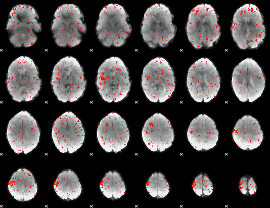 |
Component:150; Jaccard:0.047489
 |
Component:391; Jaccard:0.023801
 |
Component:391; Jaccard:0.00968
 |
Component:292; Jaccard:0.006393
 |
Correlation:0.39623
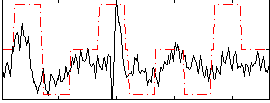 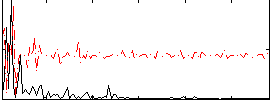 |
Correlation:0.39623
  |
Correlation:0.20491
 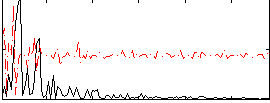 |
Correlation:-0.048596
  |
Correlation:0.3648
  |
Correlation:0.3648
  |
Correlation:-0.022653
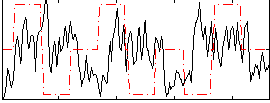 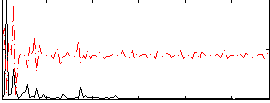 |
Component:150; Jaccard:0.070486
 |
Component:347; Jaccard:0.074287
 |
Component:150; Jaccard:0.064266
 |
Component:347; Jaccard:0.042098
 |
Component:347; Jaccard:0.020737
 |
Component:292; Jaccard:0.008871
 |
Component:182; Jaccard:0.005575
 |
Correlation:-0.048596
  |
Correlation:0.20491
  |
Correlation:-0.048596
  |
Correlation:0.20491
  |
Correlation:0.20491
  |
Correlation:-0.022653
  |
Correlation:0.03133
 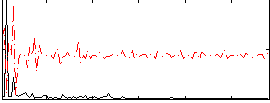 |
Component:347; Jaccard:0.069246
 |
Component:150; Jaccard:0.073139
 |
Component:337; Jaccard:0.057018
 |
Component:337; Jaccard:0.034969
 |
Component:292; Jaccard:0.015486
 |
Component:347; Jaccard:0.008549
 |
Component:301; Jaccard:0.004831
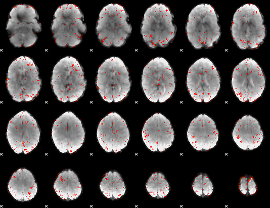 |
Correlation:0.20491
  |
Correlation:-0.048596
  |
Correlation:0.39623
  |
Correlation:0.39623
  |
Correlation:-0.022653
  |
Correlation:0.20491
  |
Correlation:0.32868
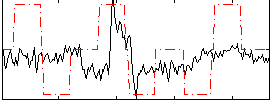  |
Component:289; Jaccard:0.067995
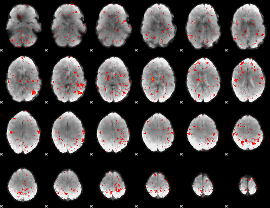 |
Component:291; Jaccard:0.065034
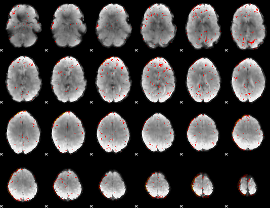 |
Component:291; Jaccard:0.046347
 |
Component:182; Jaccard:0.027813
 |
Component:182; Jaccard:0.015341
 |
Component:182; Jaccard:0.008164
 |
Component:071; Jaccard:0.004458
 |
Correlation:0.14874
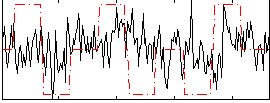 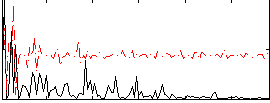 |
Correlation:0.30293
 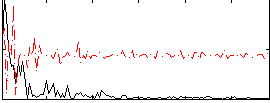 |
Correlation:0.30293
  |
Correlation:0.03133
  |
Correlation:0.03133
  |
Correlation:0.03133
  |
Correlation:-0.094618
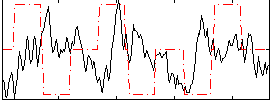  |
Component:291; Jaccard:0.0668
 |
Component:289; Jaccard:0.0632
 |
Component:182; Jaccard:0.04455
 |
Component:291; Jaccard:0.027405
 |
Component:337; Jaccard:0.013981
 |
Component:301; Jaccard:0.007127
 |
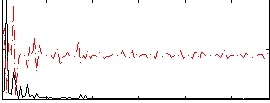
Component:381; Jaccard:0.004126
 |
Correlation:0.30293
  |
Correlation:0.14874
  |
Correlation:0.03133
  |
Correlation:0.30293
  |
Correlation:0.39623
  |
Correlation:0.32868
  |
Correlation:-0.13378
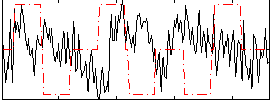 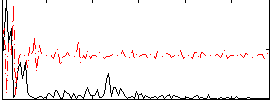 |
Component:034; Jaccard:0.065197
 |
Component:122; Jaccard:0.06051
 |
Component:289; Jaccard:0.043723
 |
Component:289; Jaccard:0.024609
 |
Component:071; Jaccard:0.011669
 |
Component:071; Jaccard:0.00683
 |
Component:391; Jaccard:0.003805
 |
Correlation:0.074081
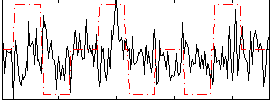  |
Correlation:0.10261
  |
Correlation:0.14874
  |
Correlation:0.14874
  |
Correlation:-0.094618
  |
Correlation:-0.094618
  |
Correlation:0.3648
  |
Component:122; Jaccard:0.064424
 |
Component:034; Jaccard:0.05805
 |
Component:122; Jaccard:0.042809
 |
Component:342; Jaccard:0.024266
 |
Component:342; Jaccard:0.010781
 |
Component:381; Jaccard:0.006576
 |
Component:031; Jaccard:0.003641
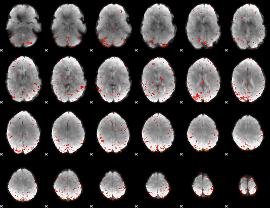 |
Correlation:0.10261
  |
Correlation:0.074081
  |
Correlation:0.10261
  |
Correlation:0.066378
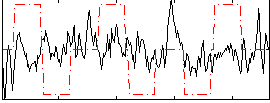 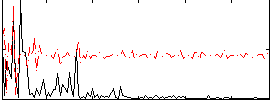 |
Correlation:0.066378
  |
Correlation:-0.13378
  |
Correlation:0.37247
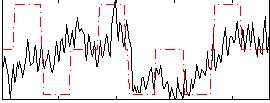  |
Component:228; Jaccard:0.062637
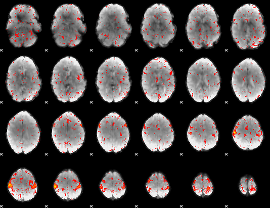 |
Component:228; Jaccard:0.057195
 |
Component:080; Jaccard:0.041052
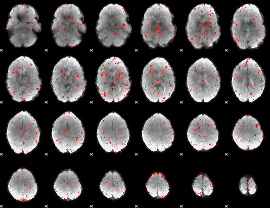 |
Component:292; Jaccard:0.023977
 |
Component:291; Jaccard:0.010698
 |
Component:031; Jaccard:0.005816
 |
Component:347; Jaccard:0.003637
 |
Correlation:-0.00081235
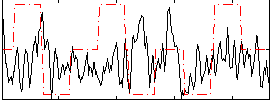 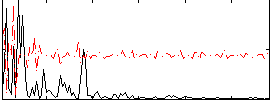 |
Correlation:-0.00081235
  |
Correlation:0.14449
  |
Correlation:-0.022653
  |
Correlation:0.30293
  |
Correlation:0.37247
  |
Correlation:0.20491
  |
Cope: 05:"neg_MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
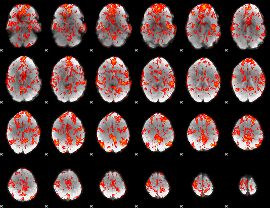 |
GLM Z-score result thresholded by 1.5
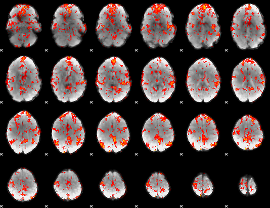 |
GLM Z-score result thresholded by 2
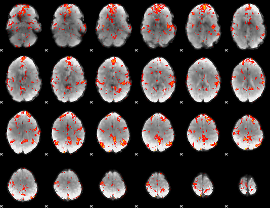 |
GLM Z-score result thresholded by 2.5
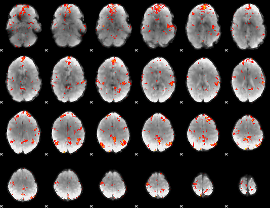 |
GLM Z-score result thresholded by 3
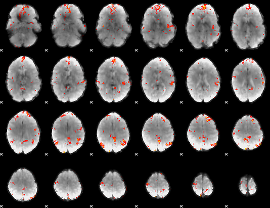 |
GLM Z-score result thresholded by 3.5
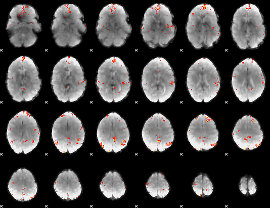 |
GLM Z-score result thresholded by 4
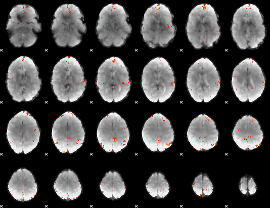 |
Component:371; Jaccard:0.11262
 |
Component:371; Jaccard:0.13572
 |
Component:149; Jaccard:0.16007
 |
Component:149; Jaccard:0.18202
 |
Component:149; Jaccard:0.1761
 |
Component:149; Jaccard:0.14696
 |
Component:149; Jaccard:0.10395
 |
Correlation:0.19096
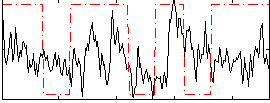  |
Correlation:0.19096
  |
Correlation:0.3237
  |
Correlation:0.3237
  |
Correlation:0.3237
  |
Correlation:0.3237
  |
Correlation:0.3237
  |
Component:149; Jaccard:0.10162
 |
Component:149; Jaccard:0.1297
 |
Component:371; Jaccard:0.15539
 |
Component:052; Jaccard:0.17026
 |
Component:052; Jaccard:0.16873
 |
Component:052; Jaccard:0.13858
 |
Component:052; Jaccard:0.093935
 |
Correlation:0.3237
  |
Correlation:0.3237
  |
Correlation:0.19096
  |
Correlation:0.16755
 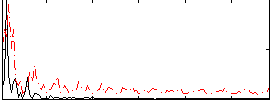 |
Correlation:0.16755
  |
Correlation:0.16755
  |
Correlation:0.16755
  |
Component:052; Jaccard:0.096013
 |
Component:052; Jaccard:0.12184
 |
Component:052; Jaccard:0.15074
 |
Component:371; Jaccard:0.16002
 |
Component:371; Jaccard:0.14201
 |
Component:371; Jaccard:0.10888
 |
Component:371; Jaccard:0.074906
 |
Correlation:0.16755
  |
Correlation:0.16755
  |
Correlation:0.16755
  |
Correlation:0.19096
  |
Correlation:0.19096
  |
Correlation:0.19096
  |
Correlation:0.19096
  |
Component:370; Jaccard:0.093989
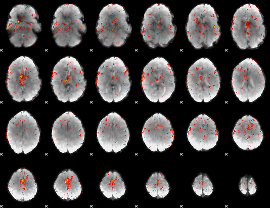 |
Component:104; Jaccard:0.1045
 |
Component:104; Jaccard:0.11753
 |
Component:104; Jaccard:0.12158
 |
Component:104; Jaccard:0.11123
 |
Component:104; Jaccard:0.08627
 |
Component:104; Jaccard:0.056747
 |
Correlation:0.16493
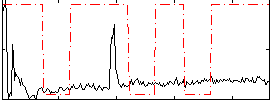 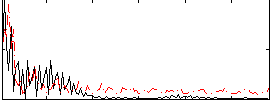 |
Correlation:-0.026764
  |
Correlation:-0.026764
  |
Correlation:-0.026764
  |
Correlation:-0.026764
  |
Correlation:-0.026764
  |
Correlation:-0.026764
  |
Component:259; Jaccard:0.091754
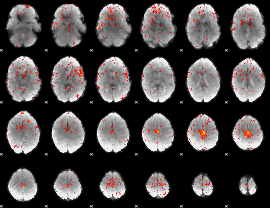 |
Component:009; Jaccard:0.096065
 |
Component:009; Jaccard:0.10024
 |
Component:250; Jaccard:0.10417
 |
Component:250; Jaccard:0.094601
 |
Component:250; Jaccard:0.075726
 |
Component:250; Jaccard:0.056115
 |
Correlation:0.10757
 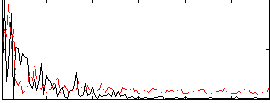 |
Correlation:0.44423
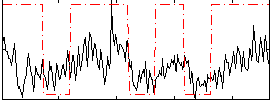  |
Correlation:0.44423
  |
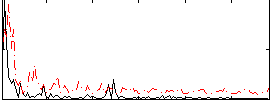
Correlation:0.10701
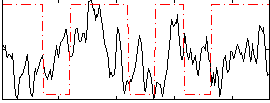  |
Correlation:0.10701
  |
Correlation:0.10701
  |
Correlation:0.10701
  |
Component:391; Jaccard:0.088217
 |
Component:370; Jaccard:0.095977
 |
Component:250; Jaccard:0.099967
 |
Component:126; Jaccard:0.099103
 |
Component:126; Jaccard:0.090623
 |
Component:126; Jaccard:0.070613
 |
Component:043; Jaccard:0.054671
 |
Correlation:0.32819
  |
Correlation:0.16493
  |
Correlation:0.10701
  |
Correlation:0.032795
  |
Correlation:0.032795
  |
Correlation:0.032795
  |
Correlation:0.34968
  |
Component:104; Jaccard:0.087389
 |
Component:259; Jaccard:0.095568
 |
Component:126; Jaccard:0.099824
 |
Component:009; Jaccard:0.093752
 |
Component:009; Jaccard:0.07875
 |
Component:009; Jaccard:0.059874
 |
Component:126; Jaccard:0.050513
 |
Correlation:-0.026764
  |
Correlation:0.10757
  |
Correlation:0.032795
  |
Correlation:0.44423
  |
Correlation:0.44423
  |
Correlation:0.44423
  |
Correlation:0.032795
  |
Component:009; Jaccard:0.085016
 |
Component:126; Jaccard:0.092581
 |
Component:259; Jaccard:0.08842
 |
Component:019; Jaccard:0.086038
 |
Component:019; Jaccard:0.078565
 |
Component:043; Jaccard:0.0591
 |
Component:009; Jaccard:0.041671
 |
Correlation:0.44423
  |
Correlation:0.032795
  |
Correlation:0.10757
  |
Correlation:0.23605
  |
Correlation:0.23605
  |
Correlation:0.34968
  |
Correlation:0.44423
  |
Component:126; Jaccard:0.081906
 |
Component:250; Jaccard:0.090824
 |
Component:391; Jaccard:0.086229
 |
Component:354; Jaccard:0.081701
 |
Component:092; Jaccard:0.071199
 |
Component:019; Jaccard:0.059049
 |
Component:019; Jaccard:0.039758
 |
Correlation:0.032795
  |
Correlation:0.10701
  |
Correlation:0.32819
  |
Correlation:0.1554
  |
Correlation:0.23609
 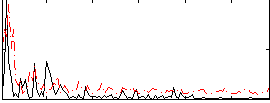 |
Correlation:0.23605
  |
Correlation:0.23605
  |
Component:246; Jaccard:0.080664
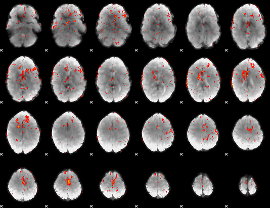 |
Component:391; Jaccard:0.090436
 |
Component:019; Jaccard:0.086158
 |
Component:092; Jaccard:0.081598
 |
Component:354; Jaccard:0.070081
 |
Component:092; Jaccard:0.054807
 |
Component:354; Jaccard:0.039002
 |
Correlation:0.089005
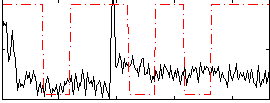 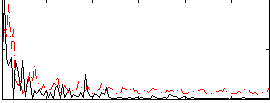 |
Correlation:0.32819
  |
Correlation:0.23605
  |
Correlation:0.23609
  |
Correlation:0.1554
  |
Correlation:0.23609
  |
Correlation:0.1554
  |
Cope: 06:"neg_REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
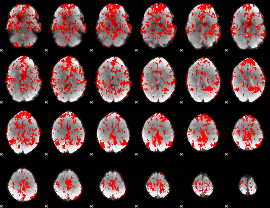 |
GLM binary result thresholded by 1.5
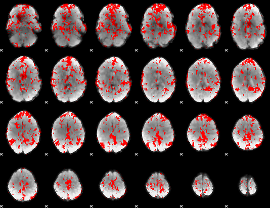 |
GLM binary result thresholded by 2
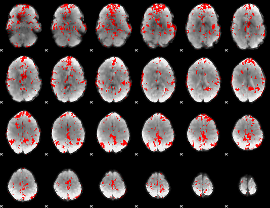 |
GLM binary result thresholded by 2.5
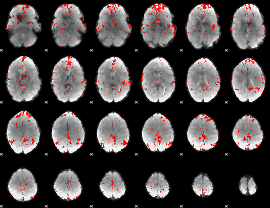 |
GLM binary result thresholded by 3
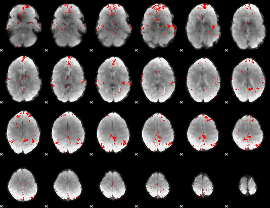 |
GLM binary result thresholded by 3.5
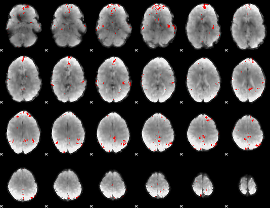 |
GLM binary result thresholded by 4
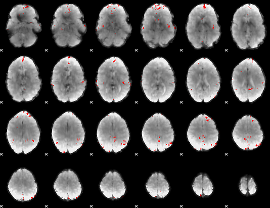 |
GLM Z-score result thresholded by 1
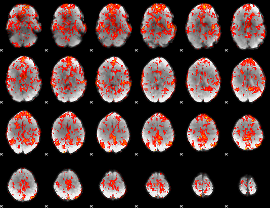 |
GLM Z-score result thresholded by 1.5
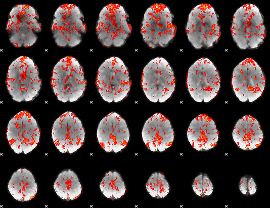 |
GLM Z-score result thresholded by 2
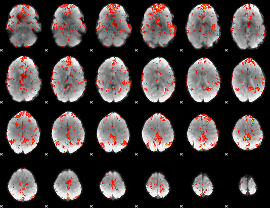 |
GLM Z-score result thresholded by 2.5
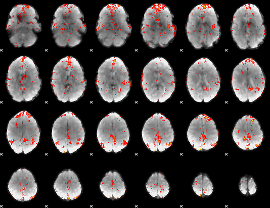 |
GLM Z-score result thresholded by 3
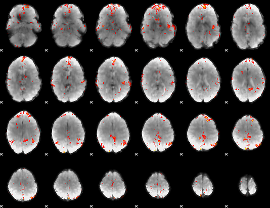 |
GLM Z-score result thresholded by 3.5
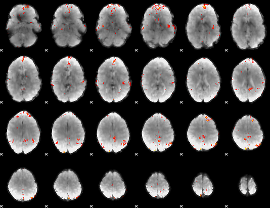 |
GLM Z-score result thresholded by 4
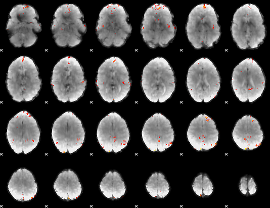 |
Component:370; Jaccard:0.10337
 |
Component:149; Jaccard:0.11915
 |
Component:149; Jaccard:0.14398
 |
Component:149; Jaccard:0.16083
 |
Component:149; Jaccard:0.15933
 |
Component:149; Jaccard:0.13131
 |
Component:149; Jaccard:0.090475
 |
Correlation:-0.0010658
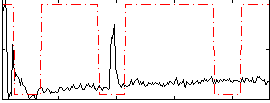 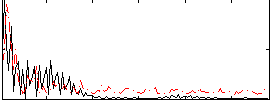 |
Correlation:0.26262
  |
Correlation:0.26262
  |
Correlation:0.26262
  |
Correlation:0.26262
  |
Correlation:0.26262
  |
Correlation:0.26262
  |
Component:149; Jaccard:0.094721
 |
Component:370; Jaccard:0.11493
 |
Component:371; Jaccard:0.11998
 |
Component:052; Jaccard:0.13189
 |
Component:052; Jaccard:0.12937
 |
Component:052; Jaccard:0.10555
 |
Component:052; Jaccard:0.072824
 |
Correlation:0.26262
  |
Correlation:-0.0010658
  |
Correlation:-0.038573
  |
Correlation:0.28457
  |
Correlation:0.28457
  |
Correlation:0.28457
  |
Correlation:0.28457
  |
Component:371; Jaccard:0.094461
 |
Component:371; Jaccard:0.10792
 |
Component:052; Jaccard:0.11941
 |
Component:371; Jaccard:0.12211
 |
Component:371; Jaccard:0.11167
 |
Component:371; Jaccard:0.087433
 |
Component:371; Jaccard:0.058834
 |
Correlation:-0.038573
  |
Correlation:-0.038573
  |
Correlation:0.28457
  |
Correlation:-0.038573
  |
Correlation:-0.038573
  |
Correlation:-0.038573
  |
Correlation:-0.038573
  |
Component:354; Jaccard:0.09424
 |
Component:354; Jaccard:0.10517
 |
Component:370; Jaccard:0.11814
 |
Component:104; Jaccard:0.11073
 |
Component:250; Jaccard:0.10284
 |
Component:250; Jaccard:0.08209
 |
Component:043; Jaccard:0.058679
 |
Correlation:0.12782
  |
Correlation:0.12782
  |
Correlation:-0.0010658
  |
Correlation:0.33548
  |
Correlation:0.22484
  |
Correlation:0.22484
  |
Correlation:-0.025341
  |
Component:259; Jaccard:0.091135
 |
Component:052; Jaccard:0.10129
 |
Component:354; Jaccard:0.11208
 |
Component:250; Jaccard:0.10897
 |
Component:104; Jaccard:0.10068
 |
Component:104; Jaccard:0.078024
 |
Component:250; Jaccard:0.056172
 |
Correlation:-0.011776
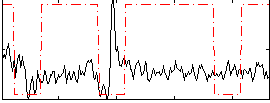 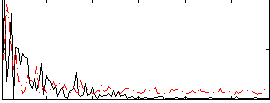 |
Correlation:0.28457
  |
Correlation:0.12782
  |
Correlation:0.22484
  |
Correlation:0.33548
  |
Correlation:0.33548
  |
Correlation:0.22484
  |
Component:010; Jaccard:0.086422
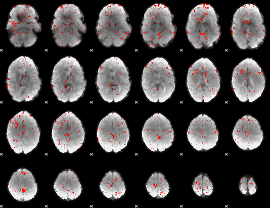 |
Component:259; Jaccard:0.097025
 |
Component:104; Jaccard:0.10638
 |
Component:354; Jaccard:0.10829
 |
Component:354; Jaccard:0.097547
 |
Component:354; Jaccard:0.073786
 |
Component:104; Jaccard:0.052566
 |
Correlation:-0.10482
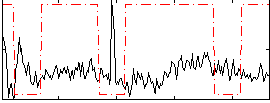  |
Correlation:-0.011776
  |
Correlation:0.33548
  |
Correlation:0.12782
  |
Correlation:0.12782
  |
Correlation:0.12782
  |
Correlation:0.33548
  |
Component:126; Jaccard:0.085611
 |
Component:126; Jaccard:0.096649
 |
Component:250; Jaccard:0.10408
 |
Component:370; Jaccard:0.10482
 |
Component:126; Jaccard:0.094199
 |
Component:126; Jaccard:0.07124
 |
Component:354; Jaccard:0.04973
 |
Correlation:0.23942
  |
Correlation:0.23942
  |
Correlation:0.22484
  |
Correlation:-0.0010658
  |
Correlation:0.23942
  |
Correlation:0.23942
  |
Correlation:0.12782
  |
Component:250; Jaccard:0.084272
 |
Component:104; Jaccard:0.096087
 |
Component:126; Jaccard:0.1039
 |
Component:126; Jaccard:0.10309
 |
Component:010; Jaccard:0.074019
 |
Component:043; Jaccard:0.057601
 |
Component:126; Jaccard:0.048801
 |
Correlation:0.22484
  |
Correlation:0.33548
  |
Correlation:0.23942
  |
Correlation:0.23942
  |
Correlation:-0.10482
  |
Correlation:-0.025341
  |
Correlation:0.23942
  |
Component:052; Jaccard:0.083818
 |
Component:010; Jaccard:0.095036
 |
Component:010; Jaccard:0.099118
 |
Component:010; Jaccard:0.092078
 |
Component:370; Jaccard:0.073688
 |
Component:010; Jaccard:0.050256
 |
Component:094; Jaccard:0.031108
 |
Correlation:0.28457
  |
Correlation:-0.10482
  |
Correlation:-0.10482
  |
Correlation:-0.10482
  |
Correlation:-0.0010658
  |
Correlation:-0.10482
  |
Correlation:0.44279
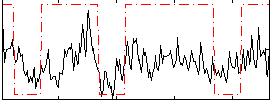 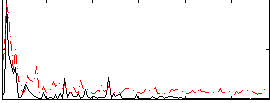 |
Component:104; Jaccard:0.082764
 |
Component:250; Jaccard:0.094797
 |
Component:259; Jaccard:0.095191
 |
Component:259; Jaccard:0.082392
 |
Component:210; Jaccard:0.067212
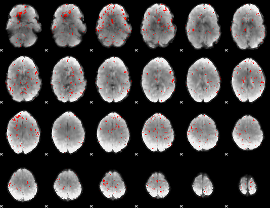 |
Component:094; Jaccard:0.049418
 |
Component:009; Jaccard:0.030958
 |
Correlation:0.33548
  |
Correlation:0.22484
  |
Correlation:-0.011776
  |
Correlation:-0.011776
  |
Correlation:0.041174
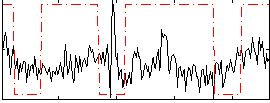  |
Correlation:0.44279
  |
Correlation:-0.038259
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































