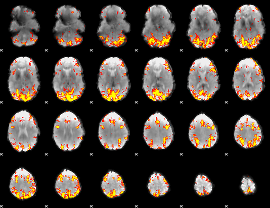
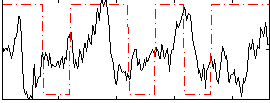
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
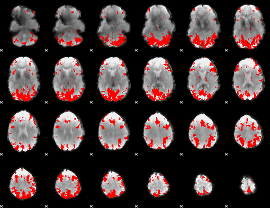
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
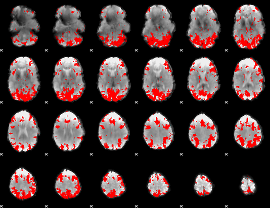 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
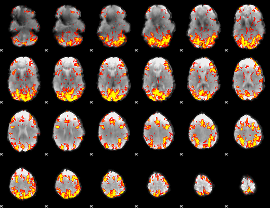 |
GLM Z-score result thresholded by 3
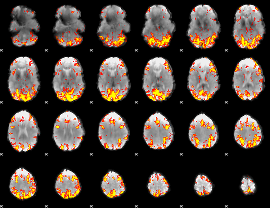 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:360; Jaccard:0.12931
 |
Component:300; Jaccard:0.15444
 |
Component:300; Jaccard:0.18711
 |
Component:300; Jaccard:0.22212
 |
Component:300; Jaccard:0.26043
 |
Component:300; Jaccard:0.29772
 |
Component:300; Jaccard:0.33199
 |
Correlation:0.33149
  |
Correlation:0.29895
  |
Correlation:0.29895
  |
Correlation:0.29895
  |
Correlation:0.29895
  |
Correlation:0.29895
  |
Correlation:0.29895
  |
Component:042; Jaccard:0.12894
 |
Component:360; Jaccard:0.15212
 |
Component:360; Jaccard:0.17571
 |
Component:360; Jaccard:0.19778
 |
Component:177; Jaccard:0.22331
 |
Component:177; Jaccard:0.25304
 |
Component:177; Jaccard:0.28143
 |
Correlation:0.14697
  |
Correlation:0.33149
  |
Correlation:0.33149
  |
Correlation:0.33149
  |
Correlation:0.32349
  |
Correlation:0.32349
  |
Correlation:0.32349
  |
Component:300; Jaccard:0.12762
 |
Component:042; Jaccard:0.14872
 |
Component:042; Jaccard:0.16964
 |
Component:177; Jaccard:0.19203
 |
Component:360; Jaccard:0.21706
 |
Component:391; Jaccard:0.24029
 |
Component:391; Jaccard:0.26057
 |
Correlation:0.29895
  |
Correlation:0.14697
  |
Correlation:0.14697
  |
Correlation:0.32349
  |
Correlation:0.33149
  |
Correlation:0.23418
  |
Correlation:0.23418
  |
Component:211; Jaccard:0.12247
 |
Component:336; Jaccard:0.13973
 |
Component:177; Jaccard:0.16213
 |
Component:042; Jaccard:0.18853
 |
Component:391; Jaccard:0.2152
 |
Component:360; Jaccard:0.22927
 |
Component:338; Jaccard:0.23497
 |
Correlation:0.014823
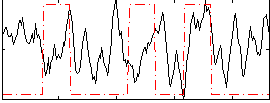  |
Correlation:0.254
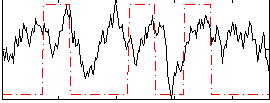 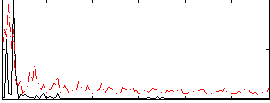 |
Correlation:0.32349
  |
Correlation:0.14697
  |
Correlation:0.23418
  |
Correlation:0.33149
  |
Correlation:0.28505
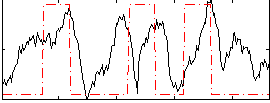  |
Component:336; Jaccard:0.12115
 |
Component:177; Jaccard:0.13479
 |
Component:391; Jaccard:0.15918
 |
Component:391; Jaccard:0.18648
 |
Component:042; Jaccard:0.20433
 |
Component:042; Jaccard:0.21301
 |
Component:360; Jaccard:0.23383
 |
Correlation:0.254
  |
Correlation:0.32349
  |
Correlation:0.23418
  |
Correlation:0.23418
  |
Correlation:0.14697
  |
Correlation:0.14697
  |
Correlation:0.33149
  |
Component:333; Jaccard:0.12109
 |
Component:271; Jaccard:0.13377
 |
Component:336; Jaccard:0.15904
 |
Component:336; Jaccard:0.17621
 |
Component:336; Jaccard:0.1905
 |
Component:338; Jaccard:0.20961
 |
Component:042; Jaccard:0.21803
 |
Correlation:0.14095
  |
Correlation:0.18969
  |
Correlation:0.254
  |
Correlation:0.254
  |
Correlation:0.254
  |
Correlation:0.28505
  |
Correlation:0.14697
  |
Component:271; Jaccard:0.11919
 |
Component:391; Jaccard:0.13333
 |
Component:271; Jaccard:0.14696
 |
Component:271; Jaccard:0.157
 |
Component:338; Jaccard:0.18276
 |
Component:336; Jaccard:0.20118
 |
Component:336; Jaccard:0.20984
 |
Correlation:0.18969
  |
Correlation:0.23418
  |
Correlation:0.18969
  |
Correlation:0.18969
  |
Correlation:0.28505
  |
Correlation:0.254
  |
Correlation:0.254
  |
Component:349; Jaccard:0.11326
 |
Component:333; Jaccard:0.13328
 |
Component:333; Jaccard:0.14425
 |
Component:338; Jaccard:0.1567
 |
Component:192; Jaccard:0.16722
 |
Component:192; Jaccard:0.1768
 |
Component:192; Jaccard:0.18154
 |
Correlation:0.19838
  |
Correlation:0.14095
  |
Correlation:0.14095
  |
Correlation:0.28505
  |
Correlation:0.3476
  |
Correlation:0.3476
  |
Correlation:0.3476
  |
Component:380; Jaccard:0.11261
 |
Component:211; Jaccard:0.1318
 |
Component:211; Jaccard:0.13992
 |
Component:192; Jaccard:0.15332
 |
Component:271; Jaccard:0.16287
 |
Component:271; Jaccard:0.16394
 |
Component:380; Jaccard:0.16151
 |
Correlation:0.1662
  |
Correlation:0.014823
  |
Correlation:0.014823
  |
Correlation:0.3476
  |
Correlation:0.18969
  |
Correlation:0.18969
  |
Correlation:0.1662
  |
Component:177; Jaccard:0.11187
 |
Component:380; Jaccard:0.12666
 |
Component:380; Jaccard:0.13964
 |
Component:333; Jaccard:0.15123
 |
Component:380; Jaccard:0.15768
 |
Component:380; Jaccard:0.16221
 |
Component:271; Jaccard:0.16081
 |
Correlation:0.32349
  |
Correlation:0.1662
  |
Correlation:0.1662
  |
Correlation:0.14095
  |
Correlation:0.1662
  |
Correlation:0.1662
  |
Correlation:0.18969
  |
Cope: 02:"REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
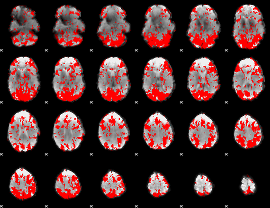 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
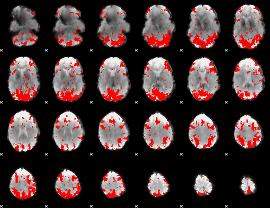 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
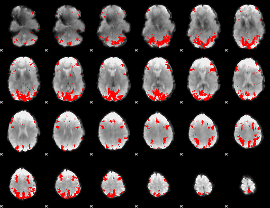 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
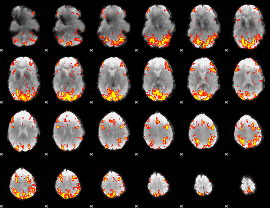 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
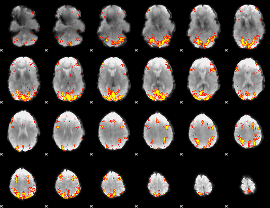 |
GLM Z-score result thresholded by 4
 |
Component:300; Jaccard:0.17043
 |
Component:300; Jaccard:0.21458
 |
Component:300; Jaccard:0.26276
 |
Component:300; Jaccard:0.30877
 |
Component:300; Jaccard:0.3484
 |
Component:300; Jaccard:0.37657
 |
Component:300; Jaccard:0.3857
 |
Correlation:0.25507
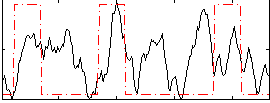 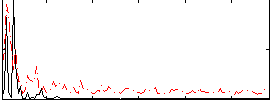 |
Correlation:0.25507
  |
Correlation:0.25507
  |
Correlation:0.25507
  |
Correlation:0.25507
  |
Correlation:0.25507
  |
Correlation:0.25507
  |
Component:042; Jaccard:0.1528
 |
Component:177; Jaccard:0.1843
 |
Component:177; Jaccard:0.22256
 |
Component:177; Jaccard:0.25873
 |
Component:177; Jaccard:0.28827
 |
Component:177; Jaccard:0.30755
 |
Component:177; Jaccard:0.3131
 |
Correlation:0.10124
  |
Correlation:0.15144
 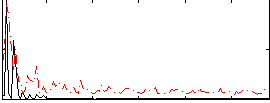 |
Correlation:0.15144
  |
Correlation:0.15144
  |
Correlation:0.15144
  |
Correlation:0.15144
  |
Correlation:0.15144
  |
Component:336; Jaccard:0.15002
 |
Component:391; Jaccard:0.17656
 |
Component:391; Jaccard:0.20916
 |
Component:391; Jaccard:0.23769
 |
Component:391; Jaccard:0.25879
 |
Component:391; Jaccard:0.2659
 |
Component:391; Jaccard:0.26055
 |
Correlation:0.30634
  |
Correlation:0.044944
  |
Correlation:0.044944
  |
Correlation:0.044944
  |
Correlation:0.044944
  |
Correlation:0.044944
  |
Correlation:0.044944
  |
Component:177; Jaccard:0.14729
 |
Component:042; Jaccard:0.17603
 |
Component:336; Jaccard:0.19548
 |
Component:336; Jaccard:0.20912
 |
Component:336; Jaccard:0.21845
 |
Component:338; Jaccard:0.23431
 |
Component:338; Jaccard:0.2485
 |
Correlation:0.15144
  |
Correlation:0.10124
  |
Correlation:0.30634
  |
Correlation:0.30634
  |
Correlation:0.30634
  |
Correlation:0.12179
  |
Correlation:0.12179
  |
Component:360; Jaccard:0.14582
 |
Component:336; Jaccard:0.17488
 |
Component:042; Jaccard:0.19316
 |
Component:042; Jaccard:0.20202
 |
Component:338; Jaccard:0.21629
 |
Component:336; Jaccard:0.21783
 |
Component:336; Jaccard:0.21026
 |
Correlation:0.089499
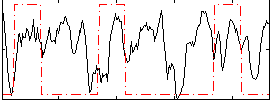  |
Correlation:0.30634
  |
Correlation:0.10124
  |
Correlation:0.10124
  |
Correlation:0.12179
  |
Correlation:0.30634
  |
Correlation:0.30634
  |
Component:391; Jaccard:0.14332
 |
Component:360; Jaccard:0.16772
 |
Component:360; Jaccard:0.18785
 |
Component:360; Jaccard:0.19893
 |
Component:042; Jaccard:0.20346
 |
Component:042; Jaccard:0.19701
 |
Component:042; Jaccard:0.18715
 |
Correlation:0.044944
  |
Correlation:0.089499
  |
Correlation:0.089499
  |
Correlation:0.089499
  |
Correlation:0.10124
  |
Correlation:0.10124
  |
Correlation:0.10124
  |
Component:333; Jaccard:0.13661
 |
Component:192; Jaccard:0.15679
 |
Component:192; Jaccard:0.17467
 |
Component:338; Jaccard:0.19354
 |
Component:360; Jaccard:0.20141
 |
Component:360; Jaccard:0.19458
 |
Component:192; Jaccard:0.18338
 |
Correlation:-0.047055
  |
Correlation:0.30382
  |
Correlation:0.30382
  |
Correlation:0.12179
  |
Correlation:0.089499
  |
Correlation:0.089499
  |
Correlation:0.30382
  |
Component:192; Jaccard:0.13635
 |
Component:333; Jaccard:0.1473
 |
Component:338; Jaccard:0.16713
 |
Component:192; Jaccard:0.18627
 |
Component:192; Jaccard:0.19236
 |
Component:192; Jaccard:0.19096
 |
Component:360; Jaccard:0.17889
 |
Correlation:0.30382
  |
Correlation:-0.047055
  |
Correlation:0.12179
  |
Correlation:0.30382
  |
Correlation:0.30382
  |
Correlation:0.30382
  |
Correlation:0.089499
  |
Component:271; Jaccard:0.12595
 |
Component:338; Jaccard:0.13988
 |
Component:333; Jaccard:0.15288
 |
Component:234; Jaccard:0.16562
 |
Component:234; Jaccard:0.17411
 |
Component:234; Jaccard:0.17819
 |
Component:234; Jaccard:0.17778
 |
Correlation:0.22319
  |
Correlation:0.12179
  |
Correlation:-0.047055
  |
Correlation:0.52923
  |
Correlation:0.52923
  |
Correlation:0.52923
  |
Correlation:0.52923
  |
Component:380; Jaccard:0.12495
 |
Component:380; Jaccard:0.13805
 |
Component:234; Jaccard:0.14973
 |
Component:380; Jaccard:0.15323
 |
Component:380; Jaccard:0.15327
 |
Component:380; Jaccard:0.14724
 |
Component:365; Jaccard:0.13696
 |
Correlation:0.15728
  |
Correlation:0.15728
  |
Correlation:0.52923
  |
Correlation:0.15728
  |
Correlation:0.15728
  |
Correlation:0.15728
  |
Correlation:0.15071
  |
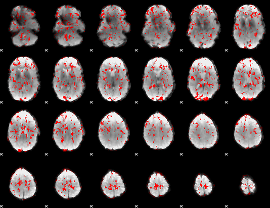
Cope: 03:"MATCH-REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:211; Jaccard:0.10107
 |
Component:211; Jaccard:0.10833
 |
Component:211; Jaccard:0.10785
 |
Component:281; Jaccard:0.097398
 |
Component:281; Jaccard:0.074293
 |
Component:281; Jaccard:0.048044
 |
Component:338; Jaccard:0.025298
 |
Correlation:-0.052547
  |
Correlation:-0.052547
  |
Correlation:-0.052547
  |
Correlation:0.10402
  |
Correlation:0.10402
  |
Correlation:0.10402
  |
Correlation:0.096765
  |
Component:349; Jaccard:0.09659
 |
Component:349; Jaccard:0.10649
 |
Component:272; Jaccard:0.10698
 |
Component:272; Jaccard:0.096725
 |
Component:272; Jaccard:0.069537
 |
Component:328; Jaccard:0.043677
 |
Component:281; Jaccard:0.024478
 |
Correlation:0.086311
  |
Correlation:0.086311
  |
Correlation:-0.041217
  |
Correlation:-0.041217
  |
Correlation:-0.041217
  |
Correlation:0.17931
  |
Correlation:0.10402
  |
Component:272; Jaccard:0.094309
 |
Component:272; Jaccard:0.10353
 |
Component:349; Jaccard:0.10577
 |
Component:105; Jaccard:0.094662
 |
Component:328; Jaccard:0.068832
 |
Component:338; Jaccard:0.04327
 |
Component:272; Jaccard:0.019798
 |
Correlation:-0.041217
  |
Correlation:-0.041217
  |
Correlation:0.086311
  |
Correlation:0.049999
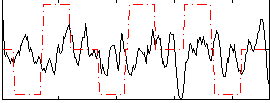  |
Correlation:0.17931
  |
Correlation:0.096765
  |
Correlation:-0.041217
  |
Component:281; Jaccard:0.089748
 |
Component:281; Jaccard:0.099188
 |
Component:281; Jaccard:0.10562
 |
Component:211; Jaccard:0.093443
 |
Component:105; Jaccard:0.067875
 |
Component:105; Jaccard:0.041279
 |
Component:105; Jaccard:0.019627
 |
Correlation:0.10402
  |
Correlation:0.10402
  |
Correlation:0.10402
  |
Correlation:-0.052547
  |
Correlation:0.049999
  |
Correlation:0.049999
  |
Correlation:0.049999
  |
Component:360; Jaccard:0.08964
 |
Component:360; Jaccard:0.098017
 |
Component:105; Jaccard:0.10298
 |
Component:349; Jaccard:0.090011
 |
Component:211; Jaccard:0.065847
 |
Component:272; Jaccard:0.040025
 |
Component:328; Jaccard:0.01884
 |
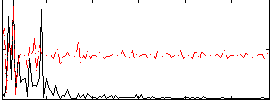
Correlation:0.14343
  |
Correlation:0.14343
  |
Correlation:0.049999
  |
Correlation:0.086311
  |
Correlation:-0.052547
  |
Correlation:-0.041217
  |
Correlation:0.17931
  |
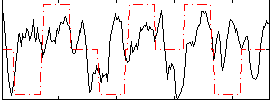
Component:105; Jaccard:0.088948
 |
Component:152; Jaccard:0.097916
 |
Component:152; Jaccard:0.098501
 |
Component:152; Jaccard:0.085582
 |
Component:383; Jaccard:0.062302
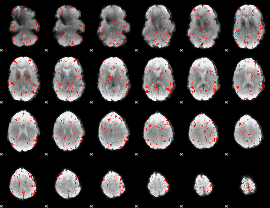 |
Component:383; Jaccard:0.039413
 |
Component:240; Jaccard:0.018648
 |
Correlation:0.049999
  |
Correlation:0.06914
  |
Correlation:0.06914
  |
Correlation:0.06914
  |
Correlation:0.19325
  |
Correlation:0.19325
  |
Correlation:0.076935
  |
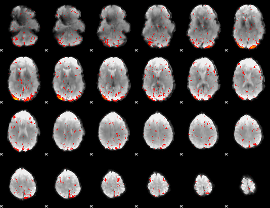
Component:152; Jaccard:0.088871
 |
Component:105; Jaccard:0.097667
 |
Component:360; Jaccard:0.095757
 |
Component:328; Jaccard:0.084161
 |
Component:349; Jaccard:0.061779
 |
Component:211; Jaccard:0.036708
 |
Component:211; Jaccard:0.01829
 |
Correlation:0.06914
  |
Correlation:0.049999
  |
Correlation:0.14343
  |
Correlation:0.17931
  |
Correlation:0.086311
  |
Correlation:-0.052547
  |
Correlation:-0.052547
  |
Component:096; Jaccard:0.080799
 |
Component:314; Jaccard:0.083448
 |
Component:328; Jaccard:0.087804
 |
Component:360; Jaccard:0.081091
 |
Component:152; Jaccard:0.060115
 |
Component:316; Jaccard:0.036604
 |
Component:223; Jaccard:0.017939
 |
Correlation:-0.046085
  |
Correlation:0.11518
  |
Correlation:0.17931
  |
Correlation:0.14343
  |
Correlation:0.06914
  |
Correlation:-0.050816
  |
Correlation:0.1714
  |
Component:347; Jaccard:0.079921
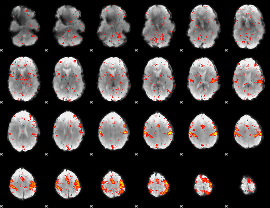 |
Component:328; Jaccard:0.082854
 |
Component:223; Jaccard:0.083381
 |
Component:383; Jaccard:0.076773
 |
Component:338; Jaccard:0.060105
 |
Component:318; Jaccard:0.036165
 |
Component:383; Jaccard:0.017593
 |
Correlation:-0.01611
  |
Correlation:0.17931
  |
Correlation:0.1714
  |
Correlation:0.19325
  |
Correlation:0.096765
  |
Correlation:0.074448
  |
Correlation:0.19325
  |
Component:314; Jaccard:0.07843
 |
Component:096; Jaccard:0.081003
 |
Component:314; Jaccard:0.081928
 |
Component:338; Jaccard:0.076256
 |
Component:318; Jaccard:0.058048
 |
Component:240; Jaccard:0.0359
 |
Component:316; Jaccard:0.016951
 |
Correlation:0.11518
  |
Correlation:-0.046085
  |
Correlation:0.11518
  |
Correlation:0.096765
  |
Correlation:0.074448
  |
Correlation:0.076935
  |
Correlation:-0.050816
  |
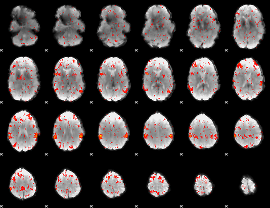
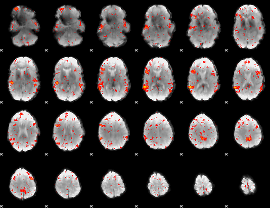
Cope: 04:"REL-MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
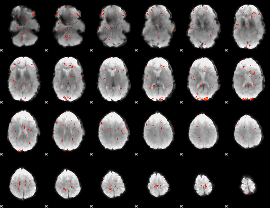 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:251; Jaccard:0.086549
 |
Component:251; Jaccard:0.084947
 |
Component:251; Jaccard:0.069747
 |
Component:251; Jaccard:0.04665
 |
Component:234; Jaccard:0.030596
 |
Component:234; Jaccard:0.01984
 |
Component:234; Jaccard:0.010177
 |
Correlation:0.15392
  |
Correlation:0.15392
  |
Correlation:0.15392
  |
Correlation:0.15392
  |
Correlation:0.39928
  |
Correlation:0.39928
  |
Correlation:0.39928
  |
Component:392; Jaccard:0.084167
 |
Component:234; Jaccard:0.068982
 |
Component:234; Jaccard:0.060214
 |
Component:234; Jaccard:0.045915
 |
Component:251; Jaccard:0.025282
 |
Component:251; Jaccard:0.013725
 |
Component:251; Jaccard:0.006826
 |
Correlation:0.26106
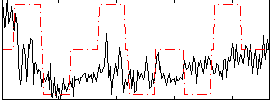  |
Correlation:0.39928
  |
Correlation:0.39928
  |
Correlation:0.39928
  |
Correlation:0.15392
  |
Correlation:0.15392
  |
Correlation:0.15392
  |
Component:204; Jaccard:0.074984
 |
Component:392; Jaccard:0.064401
 |
Component:056; Jaccard:0.043809
 |
Component:056; Jaccard:0.024178
 |
Component:285; Jaccard:0.011216
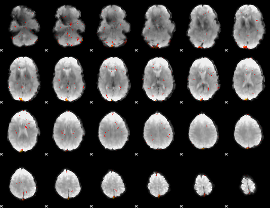 |
Component:009; Jaccard:0.00608
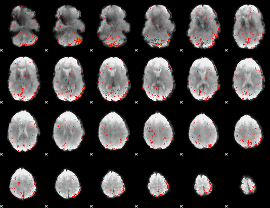 |
Component:009; Jaccard:0.004234
 |
Correlation:0.2809
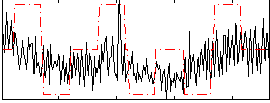  |
Correlation:0.26106
  |
Correlation:0.22789
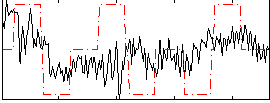  |
Correlation:0.22789
  |
Correlation:0.1361
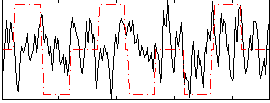  |
Correlation:0.025007
 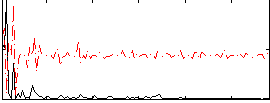 |
Correlation:0.025007
  |
Component:290; Jaccard:0.072306
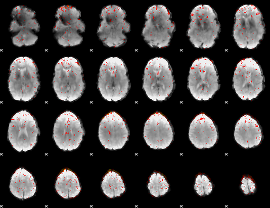 |
Component:204; Jaccard:0.062577
 |
Component:172; Jaccard:0.039884
 |
Component:218; Jaccard:0.023777
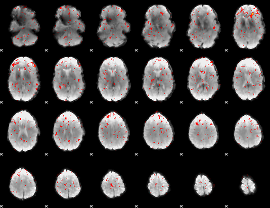 |
Component:133; Jaccard:0.010298
 |
Component:399; Jaccard:0.005964
 |
Component:399; Jaccard:0.003344
 |
Correlation:0.33342
  |
Correlation:0.2809
  |
Correlation:0.029831
 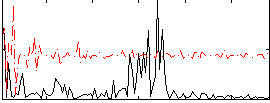 |
Correlation:0.12614
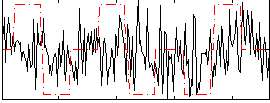  |
Correlation:0.33667
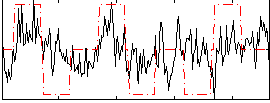  |
Correlation:0.28081
  |
Correlation:0.28081
  |
Component:375; Jaccard:0.068864
 |
Component:056; Jaccard:0.060885
 |
Component:145; Jaccard:0.039858
 |
Component:172; Jaccard:0.022461
 |
Component:307; Jaccard:0.010045
 |
Component:307; Jaccard:0.005442
 |
Component:128; Jaccard:0.003217
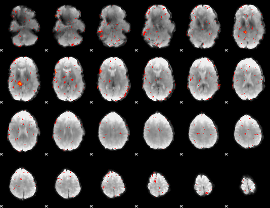 |
Correlation:0.22473
 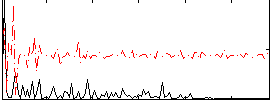 |
Correlation:0.22789
  |
Correlation:0.26077
 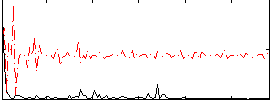 |
Correlation:0.029831
  |
Correlation:0.29234
  |
Correlation:0.29234
  |
Correlation:-0.20624
  |
Component:145; Jaccard:0.068306
 |
Component:145; Jaccard:0.059844
 |
Component:218; Jaccard:0.038581
 |
Component:285; Jaccard:0.021427
 |
Component:154; Jaccard:0.009964
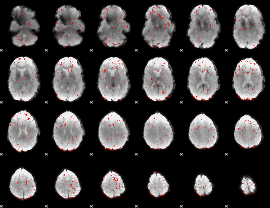 |
Component:133; Jaccard:0.005369
 |
Component:192; Jaccard:0.00311
 |
Correlation:0.26077
  |
Correlation:0.26077
  |
Correlation:0.12614
  |
Correlation:0.1361
  |
Correlation:0.25586
  |
Correlation:0.33667
  |
Correlation:-0.025948
  |
Component:234; Jaccard:0.067403
 |
Component:373; Jaccard:0.057155
 |
Component:204; Jaccard:0.03841
 |
Component:133; Jaccard:0.018472
 |
Component:172; Jaccard:0.00981
 |
Component:242; Jaccard:0.005227
 |
Component:300; Jaccard:0.002983
 |
Correlation:0.39928
  |
Correlation:0.28972
  |
Correlation:0.2809
  |
Correlation:0.33667
  |
Correlation:0.029831
  |
Correlation:0.20378
  |
Correlation:-0.026007
  |
Component:067; Jaccard:0.064606
 |
Component:375; Jaccard:0.056176
 |
Component:285; Jaccard:0.038143
 |
Component:204; Jaccard:0.018357
 |
Component:242; Jaccard:0.009434
 |
Component:154; Jaccard:0.005059
 |
Component:133; Jaccard:0.002464
 |
Correlation:0.22224
  |
Correlation:0.22473
  |
Correlation:0.1361
  |
Correlation:0.2809
  |
Correlation:0.20378
  |
Correlation:0.25586
  |
Correlation:0.33667
  |
Component:056; Jaccard:0.06391
 |
Component:290; Jaccard:0.055676
 |
Component:373; Jaccard:0.03713
 |
Component:373; Jaccard:0.018173
 |
Component:056; Jaccard:0.009421
 |
Component:192; Jaccard:0.004986
 |
Component:307; Jaccard:0.002293
 |
Correlation:0.22789
  |
Correlation:0.33342
  |
Correlation:0.28972
  |
Correlation:0.28972
  |
Correlation:0.22789
  |
Correlation:-0.025948
  |
Correlation:0.29234
  |
Component:373; Jaccard:0.063195
 |
Component:218; Jaccard:0.054462
 |
Component:392; Jaccard:0.035458
 |
Component:154; Jaccard:0.018059
 |
Component:218; Jaccard:0.009198
 |
Component:300; Jaccard:0.004971
 |
Component:285; Jaccard:0.002148
 |
Correlation:0.28972
  |
Correlation:0.12614
  |
Correlation:0.26106
  |
Correlation:0.25586
  |
Correlation:0.12614
  |
Correlation:-0.026007
  |
Correlation:0.1361
  |
Cope: 05:"neg_MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
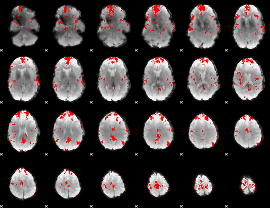
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:346; Jaccard:0.1502
 |
Component:346; Jaccard:0.17597
 |
Component:346; Jaccard:0.20209
 |
Component:346; Jaccard:0.21712
 |
Component:346; Jaccard:0.22204
 |
Component:158; Jaccard:0.22246
 |
Component:158; Jaccard:0.2084
 |
Correlation:0.19298
  |
Correlation:0.19298
  |
Correlation:0.19298
  |
Correlation:0.19298
  |
Correlation:0.19298
  |
Correlation:0.062377
  |
Correlation:0.062377
  |
Component:316; Jaccard:0.12186
 |
Component:158; Jaccard:0.14372
 |
Component:158; Jaccard:0.17347
 |
Component:158; Jaccard:0.2014
 |
Component:158; Jaccard:0.21845
 |
Component:346; Jaccard:0.20864
 |
Component:346; Jaccard:0.18359
 |
Correlation:0.12292
  |
Correlation:0.062377
  |
Correlation:0.062377
  |
Correlation:0.062377
  |
Correlation:0.062377
  |
Correlation:0.19298
  |
Correlation:0.19298
  |
Component:352; Jaccard:0.12181
 |
Component:316; Jaccard:0.14055
 |
Component:316; Jaccard:0.15914
 |
Component:316; Jaccard:0.1722
 |
Component:316; Jaccard:0.1746
 |
Component:269; Jaccard:0.1655
 |
Component:269; Jaccard:0.15228
 |
Correlation:0.2309
  |
Correlation:0.12292
  |
Correlation:0.12292
  |
Correlation:0.12292
  |
Correlation:0.12292
  |
Correlation:0.23041
  |
Correlation:0.23041
  |
Component:158; Jaccard:0.11788
 |
Component:352; Jaccard:0.13756
 |
Component:373; Jaccard:0.1502
 |
Component:373; Jaccard:0.16421
 |
Component:373; Jaccard:0.17031
 |
Component:316; Jaccard:0.1654
 |
Component:316; Jaccard:0.1487
 |
Correlation:0.062377
  |
Correlation:0.2309
  |
Correlation:0.36157
  |
Correlation:0.36157
  |
Correlation:0.36157
  |
Correlation:0.12292
  |
Correlation:0.12292
  |
Component:269; Jaccard:0.11162
 |
Component:373; Jaccard:0.13053
 |
Component:352; Jaccard:0.15013
 |
Component:269; Jaccard:0.16084
 |
Component:269; Jaccard:0.16847
 |
Component:373; Jaccard:0.16105
 |
Component:373; Jaccard:0.13947
 |
Correlation:0.23041
  |
Correlation:0.36157
  |
Correlation:0.2309
  |
Correlation:0.23041
  |
Correlation:0.23041
  |
Correlation:0.36157
  |
Correlation:0.36157
  |
Component:373; Jaccard:0.10931
 |
Component:269; Jaccard:0.12974
 |
Component:269; Jaccard:0.14701
 |
Component:352; Jaccard:0.15798
 |
Component:352; Jaccard:0.15508
 |
Component:352; Jaccard:0.1408
 |
Component:352; Jaccard:0.12411
 |
Correlation:0.36157
  |
Correlation:0.23041
  |
Correlation:0.23041
  |
Correlation:0.2309
  |
Correlation:0.2309
  |
Correlation:0.2309
  |
Correlation:0.2309
  |
Component:277; Jaccard:0.10506
 |
Component:277; Jaccard:0.11403
 |
Component:046; Jaccard:0.12475
 |
Component:046; Jaccard:0.13312
 |
Component:046; Jaccard:0.13233
 |
Component:046; Jaccard:0.12534
 |
Component:046; Jaccard:0.11059
 |
Correlation:0.078085
  |
Correlation:0.078085
  |
Correlation:0.13177
  |
Correlation:0.13177
  |
Correlation:0.13177
  |
Correlation:0.13177
  |
Correlation:0.13177
  |
Component:267; Jaccard:0.10143
 |
Component:046; Jaccard:0.11146
 |
Component:277; Jaccard:0.12331
 |
Component:277; Jaccard:0.12639
 |
Component:277; Jaccard:0.12295
 |
Component:277; Jaccard:0.11046
 |
Component:277; Jaccard:0.092371
 |
Correlation:0.14172
  |
Correlation:0.13177
  |
Correlation:0.078085
  |
Correlation:0.078085
  |
Correlation:0.078085
  |
Correlation:0.078085
  |
Correlation:0.078085
  |
Component:235; Jaccard:0.096948
 |
Component:267; Jaccard:0.10431
 |
Component:358; Jaccard:0.11116
 |
Component:358; Jaccard:0.11373
 |
Component:358; Jaccard:0.11156
 |
Component:358; Jaccard:0.10433
 |
Component:358; Jaccard:0.089608
 |
Correlation:0.090224
  |
Correlation:0.14172
  |
Correlation:0.042964
  |
Correlation:0.042964
  |
Correlation:0.042964
  |
Correlation:0.042964
  |
Correlation:0.042964
  |
Component:046; Jaccard:0.096504
 |
Component:358; Jaccard:0.10365
 |
Component:235; Jaccard:0.10571
 |
Component:106; Jaccard:0.10557
 |
Component:106; Jaccard:0.1061
 |
Component:106; Jaccard:0.097292
 |
Component:106; Jaccard:0.08689
 |
Correlation:0.13177
  |
Correlation:0.042964
  |
Correlation:0.090224
  |
Correlation:-0.042837
  |
Correlation:-0.042837
  |
Correlation:-0.042837
  |
Correlation:-0.042837
  |
Cope: 06:"neg_REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
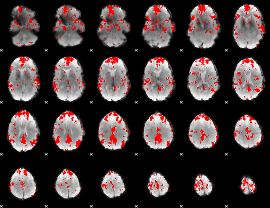 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
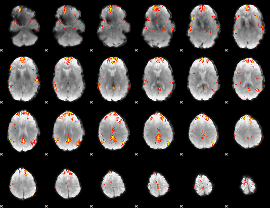
GLM Z-score result thresholded by 4
 |
Component:346; Jaccard:0.1315
 |
Component:346; Jaccard:0.15719
 |
Component:316; Jaccard:0.18963
 |
Component:316; Jaccard:0.22089
 |
Component:316; Jaccard:0.24348
 |
Component:158; Jaccard:0.26853
 |
Component:158; Jaccard:0.27448
 |
Correlation:0.013609
  |
Correlation:0.013609
  |
Correlation:0.037188
  |
Correlation:0.037188
  |
Correlation:0.037188
  |
Correlation:0.18991
  |
Correlation:0.18991
  |
Component:316; Jaccard:0.12511
 |
Component:316; Jaccard:0.15494
 |
Component:346; Jaccard:0.1851
 |
Component:346; Jaccard:0.20816
 |
Component:158; Jaccard:0.24047
 |
Component:316; Jaccard:0.2515
 |
Component:316; Jaccard:0.24554
 |
Correlation:0.037188
  |
Correlation:0.037188
  |
Correlation:0.013609
  |
Correlation:0.013609
  |
Correlation:0.18991
  |
Correlation:0.037188
  |
Correlation:0.037188
  |
Component:277; Jaccard:0.12061
 |
Component:277; Jaccard:0.14044
 |
Component:158; Jaccard:0.16786
 |
Component:158; Jaccard:0.2058
 |
Component:346; Jaccard:0.22067
 |
Component:346; Jaccard:0.22117
 |
Component:346; Jaccard:0.20431
 |
Correlation:0.26284
  |
Correlation:0.26284
  |
Correlation:0.18991
  |
Correlation:0.18991
  |
Correlation:0.013609
  |
Correlation:0.013609
  |
Correlation:0.013609
  |
Component:235; Jaccard:0.11465
 |
Component:158; Jaccard:0.13255
 |
Component:277; Jaccard:0.16042
 |
Component:277; Jaccard:0.17677
 |
Component:277; Jaccard:0.1851
 |
Component:277; Jaccard:0.18279
 |
Component:269; Jaccard:0.16878
 |
Correlation:0.070512
 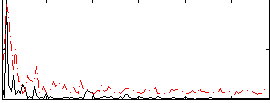 |
Correlation:0.18991
  |
Correlation:0.26284
  |
Correlation:0.26284
  |
Correlation:0.26284
  |
Correlation:0.26284
  |
Correlation:0.05587
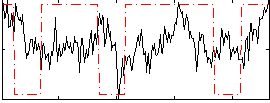 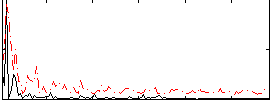 |
Component:267; Jaccard:0.10857
 |
Component:235; Jaccard:0.13136
 |
Component:235; Jaccard:0.14713
 |
Component:269; Jaccard:0.16347
 |
Component:269; Jaccard:0.1768
 |
Component:269; Jaccard:0.17627
 |
Component:277; Jaccard:0.16776
 |
Correlation:0.089833
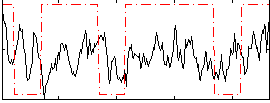  |
Correlation:0.070512
  |
Correlation:0.070512
  |
Correlation:0.05587
  |
Correlation:0.05587
  |
Correlation:0.05587
  |
Correlation:0.26284
  |
Component:158; Jaccard:0.10362
 |
Component:269; Jaccard:0.12192
 |
Component:269; Jaccard:0.14441
 |
Component:235; Jaccard:0.15527
 |
Component:235; Jaccard:0.15606
 |
Component:235; Jaccard:0.14747
 |
Component:235; Jaccard:0.13061
 |
Correlation:0.18991
  |
Correlation:0.05587
  |
Correlation:0.05587
  |
Correlation:0.070512
  |
Correlation:0.070512
  |
Correlation:0.070512
  |
Correlation:0.070512
  |
Component:269; Jaccard:0.10267
 |
Component:267; Jaccard:0.1183
 |
Component:352; Jaccard:0.12626
 |
Component:352; Jaccard:0.13326
 |
Component:352; Jaccard:0.13519
 |
Component:046; Jaccard:0.13355
 |
Component:046; Jaccard:0.12442
 |
Correlation:0.05587
  |
Correlation:0.089833
  |
Correlation:-0.058537
 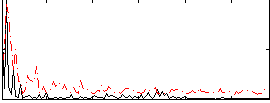 |
Correlation:-0.058537
  |
Correlation:-0.058537
  |
Correlation:0.1895
  |
Correlation:0.1895
  |
Component:352; Jaccard:0.10178
 |
Component:352; Jaccard:0.1146
 |
Component:267; Jaccard:0.12525
 |
Component:046; Jaccard:0.12644
 |
Component:046; Jaccard:0.13418
 |
Component:106; Jaccard:0.12976
 |
Component:106; Jaccard:0.12412
 |
Correlation:-0.058537
  |
Correlation:-0.058537
  |
Correlation:0.089833
  |
Correlation:0.1895
  |
Correlation:0.1895
  |
Correlation:0.3185
 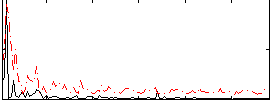 |
Correlation:0.3185
  |
Component:328; Jaccard:0.095113
 |
Component:328; Jaccard:0.10704
 |
Component:328; Jaccard:0.11833
 |
Component:106; Jaccard:0.12624
 |
Component:106; Jaccard:0.12967
 |
Component:352; Jaccard:0.12916
 |
Component:352; Jaccard:0.11762
 |
Correlation:0.1582
  |
Correlation:0.1582
  |
Correlation:0.1582
  |
Correlation:0.3185
  |
Correlation:0.3185
  |
Correlation:-0.058537
  |
Correlation:-0.058537
  |
Component:368; Jaccard:0.088783
 |
Component:106; Jaccard:0.1026
 |
Component:106; Jaccard:0.11705
 |
Component:267; Jaccard:0.12436
 |
Component:328; Jaccard:0.12387
 |
Component:328; Jaccard:0.11839
 |
Component:328; Jaccard:0.10816
 |
Correlation:0.0055402
  |
Correlation:0.3185
  |
Correlation:0.3185
  |
Correlation:0.089833
  |
Correlation:0.1582
  |
Correlation:0.1582
  |
Correlation:0.1582
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































