Task name: GAMBLING, TR=0.72, totally 253 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"PUNISH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
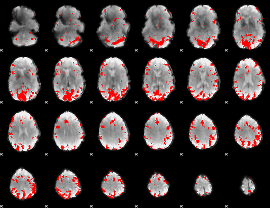 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
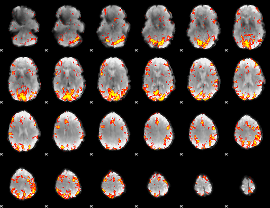 |
GLM Z-score result thresholded by 3.5
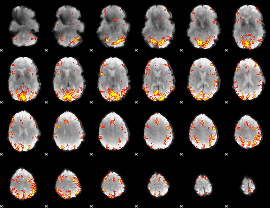 |
GLM Z-score result thresholded by 4
 |
Component:344; Jaccard:0.13126
 |
Component:344; Jaccard:0.15652
 |
Component:344; Jaccard:0.18388
 |
Component:114; Jaccard:0.22048
 |
Component:114; Jaccard:0.2602
 |
Component:114; Jaccard:0.29464
 |
Component:114; Jaccard:0.31943
 |
Correlation:0.28732
  |
Correlation:0.28732
  |
Correlation:0.28732
  |
Correlation:0.31873
  |
Correlation:0.31873
  |
Correlation:0.31873
  |
Correlation:0.31873
  |
Component:114; Jaccard:0.1176
 |
Component:114; Jaccard:0.14608
 |
Component:114; Jaccard:0.18135
 |
Component:344; Jaccard:0.21076
 |
Component:344; Jaccard:0.23256
 |
Component:344; Jaccard:0.2436
 |
Component:344; Jaccard:0.24397
 |
Correlation:0.31873
  |
Correlation:0.31873
  |
Correlation:0.31873
  |
Correlation:0.28732
  |
Correlation:0.28732
  |
Correlation:0.28732
  |
Correlation:0.28732
  |
Component:189; Jaccard:0.11617
 |
Component:189; Jaccard:0.13328
 |
Component:209; Jaccard:0.15098
 |
Component:207; Jaccard:0.1735
 |
Component:207; Jaccard:0.19482
 |
Component:207; Jaccard:0.21303
 |
Component:207; Jaccard:0.22207
 |
Correlation:0.16617
  |
Correlation:0.16617
  |
Correlation:0.29628
  |
Correlation:0.22752
  |
Correlation:0.22752
  |
Correlation:0.22752
  |
Correlation:0.22752
  |
Component:228; Jaccard:0.11581
 |
Component:209; Jaccard:0.13046
 |
Component:189; Jaccard:0.15055
 |
Component:209; Jaccard:0.16834
 |
Component:209; Jaccard:0.18007
 |
Component:209; Jaccard:0.18249
 |
Component:381; Jaccard:0.18008
 |
Correlation:0.23174
  |
Correlation:0.29628
  |
Correlation:0.16617
  |
Correlation:0.29628
  |
Correlation:0.29628
  |
Correlation:0.29628
  |
Correlation:0.32227
  |
Component:371; Jaccard:0.11538
 |
Component:040; Jaccard:0.12922
 |
Component:207; Jaccard:0.1488
 |
Component:189; Jaccard:0.16545
 |
Component:189; Jaccard:0.17429
 |
Component:381; Jaccard:0.17874
 |
Component:209; Jaccard:0.1717
 |
Correlation:0.22095
  |
Correlation:0.12714
  |
Correlation:0.22752
  |
Correlation:0.16617
  |
Correlation:0.16617
  |
Correlation:0.32227
  |
Correlation:0.29628
  |
Component:040; Jaccard:0.11181
 |
Component:055; Jaccard:0.12864
 |
Component:055; Jaccard:0.14706
 |
Component:040; Jaccard:0.16108
 |
Component:040; Jaccard:0.16973
 |
Component:189; Jaccard:0.17524
 |
Component:189; Jaccard:0.17038
 |
Correlation:0.12714
  |
Correlation:0.21156
  |
Correlation:0.21156
  |
Correlation:0.12714
  |
Correlation:0.12714
  |
Correlation:0.16617
  |
Correlation:0.16617
  |
Component:055; Jaccard:0.11086
 |
Component:371; Jaccard:0.12732
 |
Component:040; Jaccard:0.14596
 |
Component:055; Jaccard:0.161
 |
Component:381; Jaccard:0.16938
 |
Component:040; Jaccard:0.17222
 |
Component:040; Jaccard:0.1652
 |
Correlation:0.21156
  |
Correlation:0.22095
  |
Correlation:0.12714
  |
Correlation:0.21156
  |
Correlation:0.32227
  |
Correlation:0.12714
  |
Correlation:0.12714
  |
Component:224; Jaccard:0.10986
 |
Component:228; Jaccard:0.12709
 |
Component:381; Jaccard:0.13771
 |
Component:381; Jaccard:0.15552
 |
Component:055; Jaccard:0.16826
 |
Component:055; Jaccard:0.16845
 |
Component:055; Jaccard:0.16218
 |
Correlation:0.35633
  |
Correlation:0.23174
  |
Correlation:0.32227
  |
Correlation:0.32227
  |
Correlation:0.21156
  |
Correlation:0.21156
  |
Correlation:0.21156
  |
Component:209; Jaccard:0.10941
 |
Component:207; Jaccard:0.126
 |
Component:228; Jaccard:0.13658
 |
Component:061; Jaccard:0.14954
 |
Component:061; Jaccard:0.15858
 |
Component:061; Jaccard:0.16069
 |
Component:061; Jaccard:0.15434
 |
Correlation:0.29628
  |
Correlation:0.22752
  |
Correlation:0.23174
  |
Correlation:0.2332
  |
Correlation:0.2332
  |
Correlation:0.2332
  |
Correlation:0.2332
  |
Component:243; Jaccard:0.10655
 |
Component:224; Jaccard:0.12208
 |
Component:371; Jaccard:0.13496
 |
Component:228; Jaccard:0.14018
 |
Component:355; Jaccard:0.14629
 |
Component:355; Jaccard:0.15115
 |
Component:355; Jaccard:0.15052
 |
Correlation:0.13642
  |
Correlation:0.35633
  |
Correlation:0.22095
  |
Correlation:0.23174
  |
Correlation:0.10233
  |
Correlation:0.10233
  |
Correlation:0.10233
  |
Cope: 02:"REWARD" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
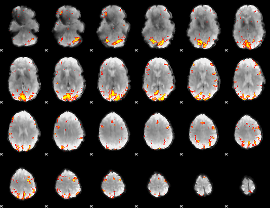
GLM Z-score result thresholded by 3.5
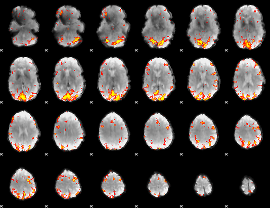 |
GLM Z-score result thresholded by 4
 |
Component:189; Jaccard:0.13063
 |
Component:114; Jaccard:0.1602
 |
Component:114; Jaccard:0.20247
 |
Component:114; Jaccard:0.24757
 |
Component:114; Jaccard:0.291
 |
Component:114; Jaccard:0.3214
 |
Component:114; Jaccard:0.33645
 |
Correlation:0.17086
  |
Correlation:0.27085
  |
Correlation:0.27085
  |
Correlation:0.27085
  |
Correlation:0.27085
  |
Correlation:0.27085
  |
Correlation:0.27085
  |
Component:344; Jaccard:0.12717
 |
Component:189; Jaccard:0.1565
 |
Component:207; Jaccard:0.18585
 |
Component:207; Jaccard:0.22134
 |
Component:207; Jaccard:0.2524
 |
Component:207; Jaccard:0.27149
 |
Component:207; Jaccard:0.27779
 |
Correlation:0.19933
  |
Correlation:0.17086
  |
Correlation:0.31277
  |
Correlation:0.31277
  |
Correlation:0.31277
  |
Correlation:0.31277
  |
Correlation:0.31277
  |
Component:114; Jaccard:0.12531
 |
Component:344; Jaccard:0.15091
 |
Component:189; Jaccard:0.17983
 |
Component:189; Jaccard:0.19815
 |
Component:344; Jaccard:0.20939
 |
Component:344; Jaccard:0.21255
 |
Component:344; Jaccard:0.20494
 |
Correlation:0.27085
  |
Correlation:0.19933
  |
Correlation:0.17086
  |
Correlation:0.17086
  |
Correlation:0.19933
  |
Correlation:0.19933
  |
Correlation:0.19933
  |
Component:207; Jaccard:0.119
 |
Component:207; Jaccard:0.15006
 |
Component:344; Jaccard:0.17487
 |
Component:344; Jaccard:0.19615
 |
Component:189; Jaccard:0.20553
 |
Component:189; Jaccard:0.19998
 |
Component:189; Jaccard:0.18338
 |
Correlation:0.31277
  |
Correlation:0.31277
  |
Correlation:0.19933
  |
Correlation:0.19933
  |
Correlation:0.17086
  |
Correlation:0.17086
  |
Correlation:0.17086
  |
Component:355; Jaccard:0.11444
 |
Component:355; Jaccard:0.1353
 |
Component:355; Jaccard:0.1571
 |
Component:355; Jaccard:0.1737
 |
Component:355; Jaccard:0.18547
 |
Component:355; Jaccard:0.18844
 |
Component:355; Jaccard:0.18154
 |
Correlation:0.34178
  |
Correlation:0.34178
  |
Correlation:0.34178
  |
Correlation:0.34178
  |
Correlation:0.34178
  |
Correlation:0.34178
  |
Correlation:0.34178
  |
Component:371; Jaccard:0.11154
 |
Component:040; Jaccard:0.12851
 |
Component:040; Jaccard:0.14567
 |
Component:040; Jaccard:0.15857
 |
Component:381; Jaccard:0.1626
 |
Component:381; Jaccard:0.16981
 |
Component:381; Jaccard:0.1646
 |
Correlation:0.25215
  |
Correlation:0.084732
  |
Correlation:0.084732
  |
Correlation:0.084732
  |
Correlation:0.13629
  |
Correlation:0.13629
  |
Correlation:0.13629
  |
Component:243; Jaccard:0.11112
 |
Component:243; Jaccard:0.12645
 |
Component:243; Jaccard:0.13927
 |
Component:061; Jaccard:0.15204
 |
Component:040; Jaccard:0.16053
 |
Component:061; Jaccard:0.15052
 |
Component:061; Jaccard:0.13542
 |
Correlation:0.13044
  |
Correlation:0.13044
  |
Correlation:0.13044
  |
Correlation:0.25275
  |
Correlation:0.084732
  |
Correlation:0.25275
  |
Correlation:0.25275
  |
Component:040; Jaccard:0.10975
 |
Component:371; Jaccard:0.12211
 |
Component:061; Jaccard:0.13794
 |
Component:381; Jaccard:0.1498
 |
Component:061; Jaccard:0.15659
 |
Component:040; Jaccard:0.15044
 |
Component:040; Jaccard:0.13336
 |
Correlation:0.084732
  |
Correlation:0.25215
  |
Correlation:0.25275
  |
Correlation:0.13629
  |
Correlation:0.25275
  |
Correlation:0.084732
  |
Correlation:0.084732
  |
Component:222; Jaccard:0.10826
 |
Component:209; Jaccard:0.12148
 |
Component:209; Jaccard:0.13737
 |
Component:209; Jaccard:0.14636
 |
Component:209; Jaccard:0.1471
 |
Component:209; Jaccard:0.13907
 |
Component:209; Jaccard:0.12317
 |
Correlation:0.17984
  |
Correlation:0.19709
  |
Correlation:0.19709
  |
Correlation:0.19709
  |
Correlation:0.19709
  |
Correlation:0.19709
  |
Correlation:0.19709
  |
Component:209; Jaccard:0.10406
 |
Component:222; Jaccard:0.12044
 |
Component:381; Jaccard:0.1344
 |
Component:243; Jaccard:0.14522
 |
Component:243; Jaccard:0.14239
 |
Component:243; Jaccard:0.13206
 |
Component:055; Jaccard:0.11896
 |
Correlation:0.19709
  |
Correlation:0.17984
  |
Correlation:0.13629
  |
Correlation:0.13044
  |
Correlation:0.13044
  |
Correlation:0.13044
  |
Correlation:0.1189
  |
Cope: 03:"PUNISH-REWARD" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:275; Jaccard:0.10292
 |
Component:275; Jaccard:0.10978
 |
Component:395; Jaccard:0.11652
 |
Component:395; Jaccard:0.1134
 |
Component:395; Jaccard:0.093035
 |
Component:395; Jaccard:0.060186
 |
Component:395; Jaccard:0.030478
 |
Correlation:0.21766
  |
Correlation:0.21766
  |
Correlation:0.46069
  |
Correlation:0.46069
  |
Correlation:0.46069
  |
Correlation:0.46069
  |
Correlation:0.46069
  |
Component:385; Jaccard:0.10178
 |
Component:329; Jaccard:0.10677
 |
Component:275; Jaccard:0.10912
 |
Component:275; Jaccard:0.093776
 |
Component:275; Jaccard:0.068291
 |
Component:028; Jaccard:0.05058
 |
Component:359; Jaccard:0.029992
 |
Correlation:0.15185
  |
Correlation:0.081158
  |
Correlation:0.21766
  |
Correlation:0.21766
  |
Correlation:0.21766
  |
Correlation:0.37933
  |
Correlation:0.4619
  |
Component:329; Jaccard:0.099271
 |
Component:385; Jaccard:0.10585
 |
Component:329; Jaccard:0.10572
 |
Component:329; Jaccard:0.091702
 |
Component:223; Jaccard:0.067779
 |
Component:359; Jaccard:0.042647
 |
Component:028; Jaccard:0.029804
 |
Correlation:0.081158
  |
Correlation:0.15185
  |
Correlation:0.081158
  |
Correlation:0.081158
  |
Correlation:0.22617
  |
Correlation:0.4619
  |
Correlation:0.37933
  |
Component:223; Jaccard:0.096
 |
Component:395; Jaccard:0.10453
 |
Component:385; Jaccard:0.10362
 |
Component:223; Jaccard:0.0896
 |
Component:028; Jaccard:0.067509
 |
Component:223; Jaccard:0.040171
 |
Component:199; Jaccard:0.02226
 |
Correlation:0.22617
  |
Correlation:0.46069
  |
Correlation:0.15185
  |
Correlation:0.22617
  |
Correlation:0.37933
  |
Correlation:0.22617
  |
Correlation:0.29578
  |
Component:395; Jaccard:0.090616
 |
Component:223; Jaccard:0.10229
 |
Component:223; Jaccard:0.10179
 |
Component:385; Jaccard:0.087302
 |
Component:329; Jaccard:0.065371
 |
Component:275; Jaccard:0.039166
 |
Component:051; Jaccard:0.021739
 |
Correlation:0.46069
  |
Correlation:0.22617
  |
Correlation:0.22617
  |
Correlation:0.15185
  |
Correlation:0.081158
  |
Correlation:0.21766
  |
Correlation:0.26638
  |
Component:224; Jaccard:0.088094
 |
Component:299; Jaccard:0.090592
 |
Component:204; Jaccard:0.086765
 |
Component:204; Jaccard:0.078674
 |
Component:204; Jaccard:0.062724
 |
Component:329; Jaccard:0.038657
 |
Component:204; Jaccard:0.021288
 |
Correlation:0.18388
 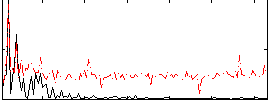 |
Correlation:0.1784
 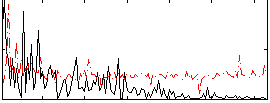 |
Correlation:0.14385
  |
Correlation:0.14385
  |
Correlation:0.14385
  |
Correlation:0.081158
  |
Correlation:0.14385
  |
Component:299; Jaccard:0.086724
 |
Component:224; Jaccard:0.087625
 |
Component:299; Jaccard:0.08556
 |
Component:028; Jaccard:0.075994
 |
Component:385; Jaccard:0.062447
 |
Component:199; Jaccard:0.038534
 |
Component:044; Jaccard:0.021248
 |
Correlation:0.1784
  |
Correlation:0.18388
  |
Correlation:0.1784
  |
Correlation:0.37933
  |
Correlation:0.15185
  |
Correlation:0.29578
  |
Correlation:0.26115
 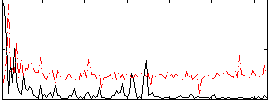 |
Component:335; Jaccard:0.084566
 |
Component:335; Jaccard:0.08665
 |
Component:335; Jaccard:0.08188
 |
Component:068; Jaccard:0.070026
 |
Component:359; Jaccard:0.054529
 |
Component:204; Jaccard:0.038249
 |
Component:217; Jaccard:0.020289
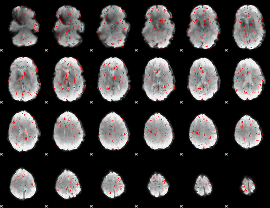 |
Correlation:0.073004
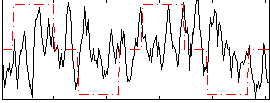  |
Correlation:0.073004
  |
Correlation:0.073004
  |
Correlation:0.11157
  |
Correlation:0.4619
  |
Correlation:0.14385
  |
Correlation:0.25725
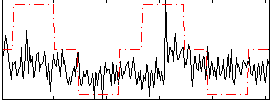 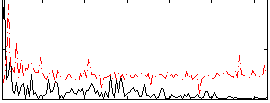 |
Component:332; Jaccard:0.084455
 |
Component:192; Jaccard:0.085243
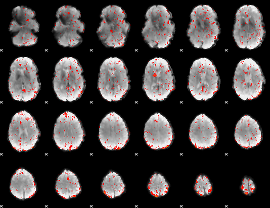 |
Component:068; Jaccard:0.079969
 |
Component:299; Jaccard:0.069558
 |
Component:199; Jaccard:0.052781
 |
Component:051; Jaccard:0.035213
 |
Component:223; Jaccard:0.019608
 |
Correlation:0.080525
  |
Correlation:0.34992
  |
Correlation:0.11157
  |
Correlation:0.1784
  |
Correlation:0.29578
  |
Correlation:0.26638
  |
Correlation:0.22617
  |
Component:192; Jaccard:0.08378
 |
Component:332; Jaccard:0.084667
 |
Component:192; Jaccard:0.079811
 |
Component:335; Jaccard:0.066716
 |
Component:044; Jaccard:0.050514
 |
Component:217; Jaccard:0.034927
 |
Component:117; Jaccard:0.019057
 |
Correlation:0.34992
  |
Correlation:0.080525
  |
Correlation:0.34992
  |
Correlation:0.073004
  |
Correlation:0.26115
  |
Correlation:0.25725
  |
Correlation:0.257
  |
Cope: 04:"neg_PUNISH" BACK
|
GLM result group-wise
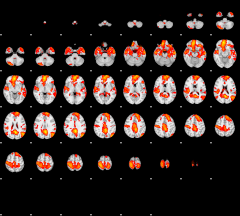 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
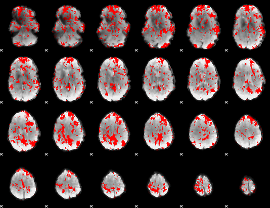 |
GLM binary result thresholded by 2
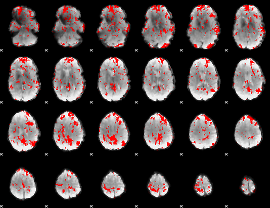 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
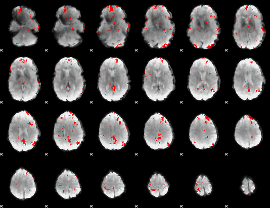 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
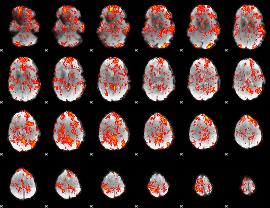 |
GLM Z-score result thresholded by 1.5
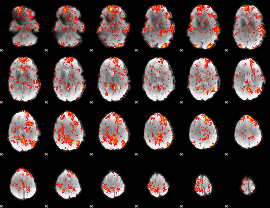 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
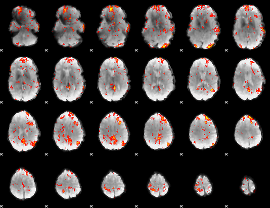 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:233; Jaccard:0.13544
 |
Component:113; Jaccard:0.16285
 |
Component:113; Jaccard:0.19265
 |
Component:113; Jaccard:0.21467
 |
Component:113; Jaccard:0.21667
 |
Component:113; Jaccard:0.1911
 |
Component:113; Jaccard:0.15232
 |
Correlation:0.3415
  |
Correlation:0.34449
  |
Correlation:0.34449
  |
Correlation:0.34449
  |
Correlation:0.34449
  |
Correlation:0.34449
  |
Correlation:0.34449
  |
Component:113; Jaccard:0.13361
 |
Component:233; Jaccard:0.16283
 |
Component:233; Jaccard:0.19051
 |
Component:233; Jaccard:0.20465
 |
Component:233; Jaccard:0.2019
 |
Component:233; Jaccard:0.17705
 |
Component:233; Jaccard:0.14293
 |
Correlation:0.34449
  |
Correlation:0.3415
  |
Correlation:0.3415
  |
Correlation:0.3415
  |
Correlation:0.3415
  |
Correlation:0.3415
  |
Correlation:0.3415
  |
Component:112; Jaccard:0.11943
 |
Component:112; Jaccard:0.13789
 |
Component:112; Jaccard:0.15346
 |
Component:112; Jaccard:0.15915
 |
Component:112; Jaccard:0.15553
 |
Component:112; Jaccard:0.13829
 |
Component:112; Jaccard:0.11321
 |
Correlation:0.25378
  |
Correlation:0.25378
  |
Correlation:0.25378
  |
Correlation:0.25378
  |
Correlation:0.25378
  |
Correlation:0.25378
  |
Correlation:0.25378
  |
Component:160; Jaccard:0.10682
 |
Component:160; Jaccard:0.11483
 |
Component:160; Jaccard:0.11812
 |
Component:160; Jaccard:0.11467
 |
Component:160; Jaccard:0.10375
 |
Component:160; Jaccard:0.086756
 |
Component:160; Jaccard:0.065177
 |
Correlation:0.20678
  |
Correlation:0.20678
  |
Correlation:0.20678
  |
Correlation:0.20678
  |
Correlation:0.20678
  |
Correlation:0.20678
  |
Correlation:0.20678
  |
Component:023; Jaccard:0.098721
 |
Component:023; Jaccard:0.10753
 |
Component:023; Jaccard:0.11227
 |
Component:373; Jaccard:0.11042
 |
Component:373; Jaccard:0.10194
 |
Component:373; Jaccard:0.081189
 |
Component:337; Jaccard:0.063206
 |
Correlation:0.16899
  |
Correlation:0.16899
  |
Correlation:0.16899
  |
Correlation:0.24031
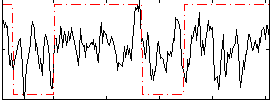  |
Correlation:0.24031
  |
Correlation:0.24031
  |
Correlation:0.092607
 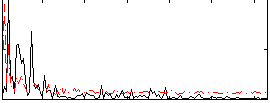 |
Component:337; Jaccard:0.095202
 |
Component:337; Jaccard:0.10211
 |
Component:373; Jaccard:0.10951
 |
Component:023; Jaccard:0.10921
 |
Component:023; Jaccard:0.096206
 |
Component:337; Jaccard:0.078932
 |
Component:270; Jaccard:0.06196
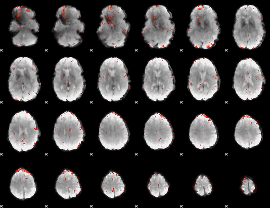 |
Correlation:0.092607
  |
Correlation:0.092607
  |
Correlation:0.24031
  |
Correlation:0.16899
  |
Correlation:0.16899
  |
Correlation:0.092607
  |
Correlation:0.3404
  |
Component:101; Jaccard:0.094596
 |
Component:101; Jaccard:0.10075
 |
Component:337; Jaccard:0.10544
 |
Component:337; Jaccard:0.10266
 |
Component:337; Jaccard:0.094345
 |
Component:023; Jaccard:0.078169
 |
Component:023; Jaccard:0.059229
 |
Correlation:0.06083
  |
Correlation:0.06083
  |
Correlation:0.092607
  |
Correlation:0.092607
  |
Correlation:0.092607
  |
Correlation:0.16899
  |
Correlation:0.16899
  |
Component:378; Jaccard:0.088517
 |
Component:373; Jaccard:0.098775
 |
Component:101; Jaccard:0.1044
 |
Component:101; Jaccard:0.10041
 |
Component:101; Jaccard:0.089305
 |
Component:101; Jaccard:0.073098
 |
Component:373; Jaccard:0.057836
 |
Correlation:0.092274
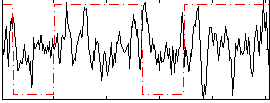  |
Correlation:0.24031
  |
Correlation:0.06083
  |
Correlation:0.06083
  |
Correlation:0.06083
  |
Correlation:0.06083
  |
Correlation:0.24031
  |
Component:373; Jaccard:0.087461
 |
Component:324; Jaccard:0.094595
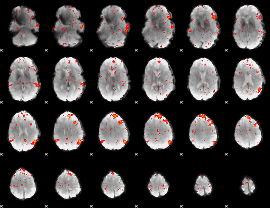 |
Component:324; Jaccard:0.09911
 |
Component:324; Jaccard:0.097614
 |
Component:324; Jaccard:0.086501
 |
Component:270; Jaccard:0.07235
 |
Component:101; Jaccard:0.053976
 |
Correlation:0.24031
  |
Correlation:0.26021
  |
Correlation:0.26021
  |
Correlation:0.26021
  |
Correlation:0.26021
  |
Correlation:0.3404
  |
Correlation:0.06083
  |
Component:293; Jaccard:0.086785
 |
Component:293; Jaccard:0.093779
 |
Component:293; Jaccard:0.095405
 |
Component:293; Jaccard:0.093438
 |
Component:293; Jaccard:0.083138
 |
Component:134; Jaccard:0.069702
 |
Component:134; Jaccard:0.052978
 |
Correlation:0.17791
 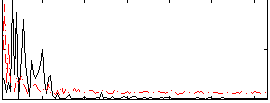 |
Correlation:0.17791
  |
Correlation:0.17791
  |
Correlation:0.17791
  |
Correlation:0.17791
  |
Correlation:0.18117
  |
Correlation:0.18117
  |
Cope: 05:"neg_REWARD" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
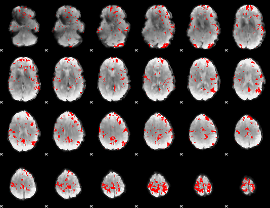 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
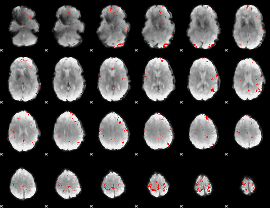 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
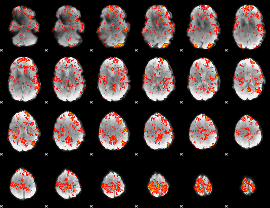 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
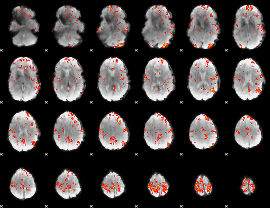 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
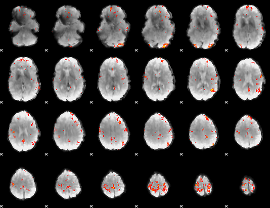 |
GLM Z-score result thresholded by 4
 |
Component:329; Jaccard:0.13302
 |
Component:329; Jaccard:0.15001
 |
Component:329; Jaccard:0.15946
 |
Component:112; Jaccard:0.16218
 |
Component:112; Jaccard:0.15087
 |
Component:112; Jaccard:0.12906
 |
Component:112; Jaccard:0.098319
 |
Correlation:0.11939
 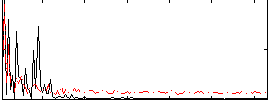 |
Correlation:0.11939
  |
Correlation:0.11939
  |
Correlation:0.20445
  |
Correlation:0.20445
  |
Correlation:0.20445
  |
Correlation:0.20445
  |
Component:223; Jaccard:0.13049
 |
Component:223; Jaccard:0.14685
 |
Component:223; Jaccard:0.15736
 |
Component:329; Jaccard:0.15366
 |
Component:329; Jaccard:0.1362
 |
Component:113; Jaccard:0.11173
 |
Component:113; Jaccard:0.075536
 |
Correlation:0.3465
  |
Correlation:0.3465
  |
Correlation:0.3465
  |
Correlation:0.11939
  |
Correlation:0.11939
  |
Correlation:0.064527
  |
Correlation:0.064527
  |
Component:112; Jaccard:0.12044
 |
Component:113; Jaccard:0.14009
 |
Component:112; Jaccard:0.1545
 |
Component:223; Jaccard:0.15187
 |
Component:113; Jaccard:0.13509
 |
Component:329; Jaccard:0.10839
 |
Component:329; Jaccard:0.075437
 |
Correlation:0.20445
  |
Correlation:0.064527
  |
Correlation:0.20445
  |
Correlation:0.3465
  |
Correlation:0.064527
  |
Correlation:0.11939
  |
Correlation:0.11939
  |
Component:113; Jaccard:0.11936
 |
Component:112; Jaccard:0.13989
 |
Component:113; Jaccard:0.15286
 |
Component:113; Jaccard:0.15101
 |
Component:223; Jaccard:0.13447
 |
Component:223; Jaccard:0.10659
 |
Component:223; Jaccard:0.07432
 |
Correlation:0.064527
  |
Correlation:0.20445
  |
Correlation:0.064527
  |
Correlation:0.064527
  |
Correlation:0.3465
  |
Correlation:0.3465
  |
Correlation:0.3465
  |
Component:101; Jaccard:0.11769
 |
Component:101; Jaccard:0.13154
 |
Component:101; Jaccard:0.14126
 |
Component:101; Jaccard:0.13656
 |
Component:101; Jaccard:0.12194
 |
Component:101; Jaccard:0.098223
 |
Component:101; Jaccard:0.070815
 |
Correlation:0.011421
  |
Correlation:0.011421
  |
Correlation:0.011421
  |
Correlation:0.011421
  |
Correlation:0.011421
  |
Correlation:0.011421
  |
Correlation:0.011421
  |
Component:233; Jaccard:0.11288
 |
Component:233; Jaccard:0.12817
 |
Component:233; Jaccard:0.13604
 |
Component:233; Jaccard:0.13294
 |
Component:233; Jaccard:0.11649
 |
Component:233; Jaccard:0.090804
 |
Component:233; Jaccard:0.061588
 |
Correlation:0.11046
  |
Correlation:0.11046
  |
Correlation:0.11046
  |
Correlation:0.11046
  |
Correlation:0.11046
  |
Correlation:0.11046
  |
Correlation:0.11046
  |
Component:325; Jaccard:0.10833
 |
Component:325; Jaccard:0.11028
 |
Component:396; Jaccard:0.1092
 |
Component:396; Jaccard:0.10472
 |
Component:396; Jaccard:0.093696
 |
Component:396; Jaccard:0.076469
 |
Component:396; Jaccard:0.054634
 |
Correlation:0.082484
  |
Correlation:0.082484
  |
Correlation:0.020152
  |
Correlation:0.020152
  |
Correlation:0.020152
  |
Correlation:0.020152
  |
Correlation:0.020152
  |
Component:160; Jaccard:0.099965
 |
Component:396; Jaccard:0.10559
 |
Component:325; Jaccard:0.10717
 |
Component:325; Jaccard:0.096846
 |
Component:160; Jaccard:0.0834
 |
Component:160; Jaccard:0.064026
 |
Component:023; Jaccard:0.044428
 |
Correlation:0.092223
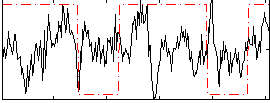  |
Correlation:0.020152
  |
Correlation:0.082484
  |
Correlation:0.082484
  |
Correlation:0.092223
  |
Correlation:0.092223
  |
Correlation:0.015682
  |
Component:396; Jaccard:0.099087
 |
Component:160; Jaccard:0.10481
 |
Component:160; Jaccard:0.10413
 |
Component:160; Jaccard:0.09682
 |
Component:325; Jaccard:0.083296
 |
Component:325; Jaccard:0.063869
 |
Component:160; Jaccard:0.044097
 |
Correlation:0.020152
  |
Correlation:0.092223
  |
Correlation:0.092223
  |
Correlation:0.092223
  |
Correlation:0.082484
  |
Correlation:0.082484
  |
Correlation:0.092223
  |
Component:378; Jaccard:0.096358
 |
Component:378; Jaccard:0.099507
 |
Component:074; Jaccard:0.097043
 |
Component:373; Jaccard:0.090624
 |
Component:373; Jaccard:0.076341
 |
Component:023; Jaccard:0.059941
 |
Component:325; Jaccard:0.043352
 |
Correlation:0.017016
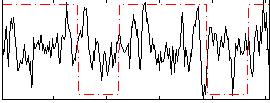  |
Correlation:0.017016
  |
Correlation:0.15991
 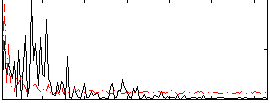 |
Correlation:0.080612
  |
Correlation:0.080612
  |
Correlation:0.015682
  |
Correlation:0.082484
  |
Cope: 06:"REWARD-PUNISH" BACK
|
GLM result group-wise
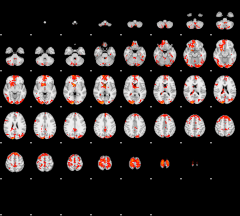 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:319; Jaccard:0.11307
 |
Component:319; Jaccard:0.13795
 |
Component:319; Jaccard:0.15457
 |
Component:319; Jaccard:0.15438
 |
Component:319; Jaccard:0.13732
 |
Component:319; Jaccard:0.11036
 |
Component:319; Jaccard:0.081008
 |
Correlation:0.69705
  |
Correlation:0.69705
  |
Correlation:0.69705
  |
Correlation:0.69705
  |
Correlation:0.69705
  |
Correlation:0.69705
  |
Correlation:0.69705
  |
Component:251; Jaccard:0.10209
 |
Component:251; Jaccard:0.11691
 |
Component:251; Jaccard:0.12156
 |
Component:251; Jaccard:0.1156
 |
Component:336; Jaccard:0.1052
 |
Component:336; Jaccard:0.088199
 |
Component:336; Jaccard:0.064174
 |
Correlation:0.39205
  |
Correlation:0.39205
  |
Correlation:0.39205
  |
Correlation:0.39205
  |
Correlation:0.27592
  |
Correlation:0.27592
  |
Correlation:0.27592
  |
Component:309; Jaccard:0.090982
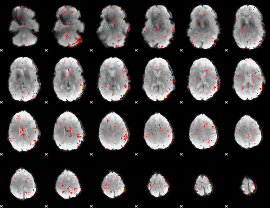 |
Component:309; Jaccard:0.10285
 |
Component:309; Jaccard:0.10823
 |
Component:336; Jaccard:0.1151
 |
Component:251; Jaccard:0.096478
 |
Component:251; Jaccard:0.073744
 |
Component:251; Jaccard:0.053836
 |
Correlation:0.49421
 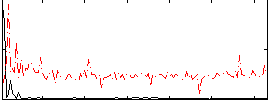 |
Correlation:0.49421
  |
Correlation:0.49421
  |
Correlation:0.27592
  |
Correlation:0.39205
  |
Correlation:0.39205
  |
Correlation:0.39205
  |
Component:266; Jaccard:0.090255
 |
Component:336; Jaccard:0.092432
 |
Component:336; Jaccard:0.10712
 |
Component:309; Jaccard:0.10244
 |
Component:309; Jaccard:0.086267
 |
Component:309; Jaccard:0.06476
 |
Component:309; Jaccard:0.044194
 |
Correlation:0.22282
  |
Correlation:0.27592
  |
Correlation:0.27592
  |
Correlation:0.49421
  |
Correlation:0.49421
  |
Correlation:0.49421
  |
Correlation:0.49421
  |
Component:337; Jaccard:0.087375
 |
Component:266; Jaccard:0.090179
 |
Component:266; Jaccard:0.083089
 |
Component:156; Jaccard:0.078095
 |
Component:270; Jaccard:0.070068
 |
Component:270; Jaccard:0.052452
 |
Component:270; Jaccard:0.038848
 |
Correlation:0.1313
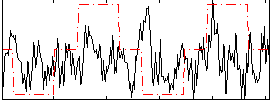  |
Correlation:0.22282
  |
Correlation:0.22282
  |
Correlation:0.18915
  |
Correlation:0.29621
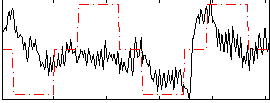  |
Correlation:0.29621
  |
Correlation:0.29621
  |
Component:318; Jaccard:0.08432
 |
Component:337; Jaccard:0.08677
 |
Component:156; Jaccard:0.081089
 |
Component:270; Jaccard:0.074336
 |
Component:156; Jaccard:0.063794
 |
Component:156; Jaccard:0.047765
 |
Component:156; Jaccard:0.031343
 |
Correlation:0.30821
  |
Correlation:0.1313
  |
Correlation:0.18915
  |
Correlation:0.29621
  |
Correlation:0.18915
  |
Correlation:0.18915
  |
Correlation:0.18915
  |
Component:128; Jaccard:0.076919
 |
Component:318; Jaccard:0.08223
 |
Component:337; Jaccard:0.077562
 |
Component:134; Jaccard:0.068707
 |
Component:134; Jaccard:0.056364
 |
Component:276; Jaccard:0.043178
 |
Component:006; Jaccard:0.030622
 |
Correlation:0.19454
  |
Correlation:0.30821
  |
Correlation:0.1313
  |
Correlation:0.17318
  |
Correlation:0.17318
  |
Correlation:0.29954
  |
Correlation:0.094708
  |
Component:336; Jaccard:0.075925
 |
Component:156; Jaccard:0.077512
 |
Component:134; Jaccard:0.076821
 |
Component:266; Jaccard:0.066618
 |
Component:276; Jaccard:0.053659
 |
Component:006; Jaccard:0.042994
 |
Component:134; Jaccard:0.030263
 |
Correlation:0.27592
  |
Correlation:0.18915
  |
Correlation:0.17318
  |
Correlation:0.22282
  |
Correlation:0.29954
  |
Correlation:0.094708
  |
Correlation:0.17318
  |
Component:038; Jaccard:0.074015
 |
Component:128; Jaccard:0.075793
 |
Component:318; Jaccard:0.074416
 |
Component:337; Jaccard:0.065249
 |
Component:006; Jaccard:0.053341
 |
Component:134; Jaccard:0.042046
 |
Component:037; Jaccard:0.030001
 |
Correlation:0.24089
  |
Correlation:0.19454
  |
Correlation:0.30821
  |
Correlation:0.1313
  |
Correlation:0.094708
  |
Correlation:0.17318
  |
Correlation:0.19738
  |
Component:233; Jaccard:0.073823
 |
Component:134; Jaccard:0.075502
 |
Component:270; Jaccard:0.071449
 |
Component:276; Jaccard:0.065107
 |
Component:037; Jaccard:0.051159
 |
Component:037; Jaccard:0.039345
 |
Component:276; Jaccard:0.029141
 |
Correlation:0.13587
  |
Correlation:0.17318
  |
Correlation:0.29621
  |
Correlation:0.29954
  |
Correlation:0.19738
  |
Correlation:0.19738
  |
Correlation:0.29954
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































