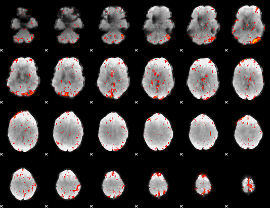
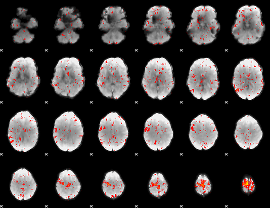
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"MATCH" BACK
|
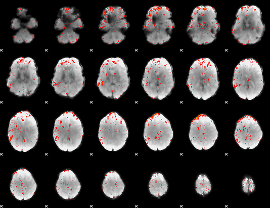
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
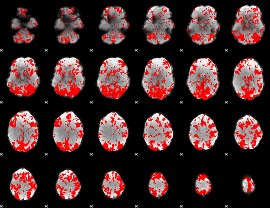 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
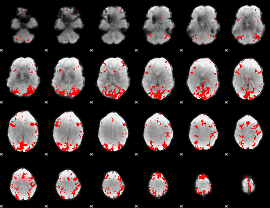 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
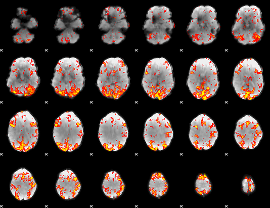 |
GLM Z-score result thresholded by 3
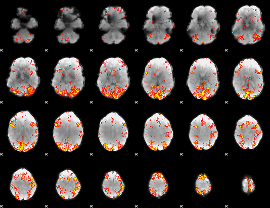 |
GLM Z-score result thresholded by 3.5
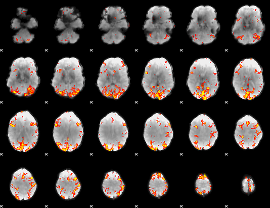 |
GLM Z-score result thresholded by 4
 |
Component:197; Jaccard:0.12418
 |
Component:127; Jaccard:0.14392
 |
Component:127; Jaccard:0.18267
 |
Component:127; Jaccard:0.22736
 |
Component:127; Jaccard:0.26748
 |
Component:127; Jaccard:0.2968
 |
Component:127; Jaccard:0.30368
 |
Correlation:0.27649
  |
Correlation:0.35558
  |
Correlation:0.35558
  |
Correlation:0.35558
  |
Correlation:0.35558
  |
Correlation:0.35558
  |
Correlation:0.35558
  |
Component:127; Jaccard:0.11193
 |
Component:197; Jaccard:0.14343
 |
Component:371; Jaccard:0.17397
 |
Component:371; Jaccard:0.21191
 |
Component:371; Jaccard:0.2506
 |
Component:371; Jaccard:0.27326
 |
Component:371; Jaccard:0.27723
 |
Correlation:0.35558
  |
Correlation:0.27649
  |
Correlation:0.16407
  |
Correlation:0.16407
  |
Correlation:0.16407
  |
Correlation:0.16407
  |
Correlation:0.16407
  |
Component:371; Jaccard:0.11081
 |
Component:371; Jaccard:0.13908
 |
Component:337; Jaccard:0.16168
 |
Component:337; Jaccard:0.19007
 |
Component:337; Jaccard:0.21236
 |
Component:179; Jaccard:0.23527
 |
Component:179; Jaccard:0.24918
 |
Correlation:0.16407
  |
Correlation:0.16407
  |
Correlation:0.37419
  |
Correlation:0.37419
  |
Correlation:0.37419
  |
Correlation:0.22976
  |
Correlation:0.22976
  |
Component:337; Jaccard:0.10795
 |
Component:337; Jaccard:0.13285
 |
Component:197; Jaccard:0.16148
 |
Component:179; Jaccard:0.17448
 |
Component:179; Jaccard:0.20686
 |
Component:337; Jaccard:0.22488
 |
Component:337; Jaccard:0.22325
 |
Correlation:0.37419
  |
Correlation:0.37419
  |
Correlation:0.27649
  |
Correlation:0.22976
  |
Correlation:0.22976
  |
Correlation:0.37419
  |
Correlation:0.37419
  |
Component:243; Jaccard:0.10381
 |
Component:270; Jaccard:0.12426
 |
Component:255; Jaccard:0.14679
 |
Component:197; Jaccard:0.17337
 |
Component:395; Jaccard:0.18078
 |
Component:395; Jaccard:0.19302
 |
Component:395; Jaccard:0.19679
 |
Correlation:0.20006
  |
Correlation:0.19262
  |
Correlation:0.31957
  |
Correlation:0.27649
  |
Correlation:0.23449
  |
Correlation:0.23449
  |
Correlation:0.23449
  |
Component:270; Jaccard:0.10297
 |
Component:255; Jaccard:0.12361
 |
Component:270; Jaccard:0.14541
 |
Component:255; Jaccard:0.16608
 |
Component:255; Jaccard:0.17899
 |
Component:270; Jaccard:0.18325
 |
Component:270; Jaccard:0.17344
 |
Correlation:0.19262
  |
Correlation:0.31957
  |
Correlation:0.19262
  |
Correlation:0.31957
  |
Correlation:0.31957
  |
Correlation:0.19262
  |
Correlation:0.19262
  |
Component:255; Jaccard:0.10291
 |
Component:131; Jaccard:0.12071
 |
Component:179; Jaccard:0.14068
 |
Component:270; Jaccard:0.16485
 |
Component:270; Jaccard:0.17767
 |
Component:255; Jaccard:0.18057
 |
Component:255; Jaccard:0.17175
 |
Correlation:0.31957
  |
Correlation:0.31973
  |
Correlation:0.22976
  |
Correlation:0.19262
  |
Correlation:0.19262
  |
Correlation:0.31957
  |
Correlation:0.31957
  |
Component:131; Jaccard:0.10205
 |
Component:395; Jaccard:0.11685
 |
Component:395; Jaccard:0.13896
 |
Component:395; Jaccard:0.16104
 |
Component:197; Jaccard:0.17616
 |
Component:197; Jaccard:0.16925
 |
Component:066; Jaccard:0.16103
 |
Correlation:0.31973
  |
Correlation:0.23449
  |
Correlation:0.23449
  |
Correlation:0.23449
  |
Correlation:0.27649
  |
Correlation:0.27649
  |
Correlation:0.11692
 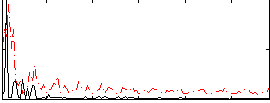 |
Component:119; Jaccard:0.099151
 |
Component:243; Jaccard:0.11365
 |
Component:131; Jaccard:0.13823
 |
Component:131; Jaccard:0.15152
 |
Component:131; Jaccard:0.15997
 |
Component:066; Jaccard:0.16206
 |
Component:197; Jaccard:0.15681
 |
Correlation:0.17277
 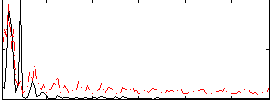 |
Correlation:0.20006
  |
Correlation:0.31973
  |
Correlation:0.31973
  |
Correlation:0.31973
  |
Correlation:0.11692
  |
Correlation:0.27649
  |
Component:096; Jaccard:0.096782
 |
Component:179; Jaccard:0.11233
 |
Component:066; Jaccard:0.1297
 |
Component:066; Jaccard:0.14704
 |
Component:066; Jaccard:0.15809
 |
Component:131; Jaccard:0.15854
 |
Component:131; Jaccard:0.15099
 |
Correlation:0.26821
  |
Correlation:0.22976
  |
Correlation:0.11692
  |
Correlation:0.11692
  |
Correlation:0.11692
  |
Correlation:0.31973
  |
Correlation:0.31973
  |
Cope: 02:"REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
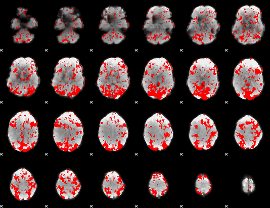 |
GLM binary result thresholded by 1.5
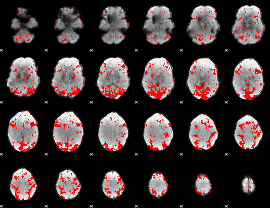 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
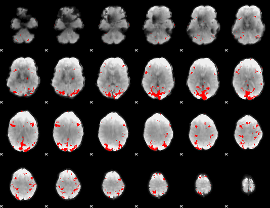 |
GLM binary result thresholded by 3.5
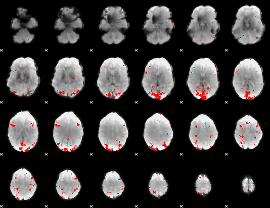 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
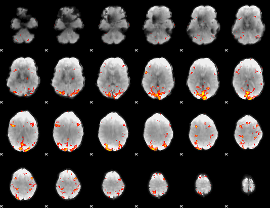 |
GLM Z-score result thresholded by 3.5
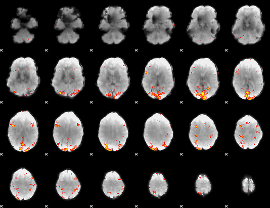 |
GLM Z-score result thresholded by 4
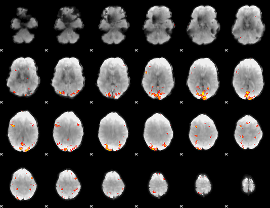 |
Component:371; Jaccard:0.16583
 |
Component:371; Jaccard:0.21313
 |
Component:179; Jaccard:0.26012
 |
Component:179; Jaccard:0.29682
 |
Component:179; Jaccard:0.31087
 |
Component:179; Jaccard:0.29237
 |
Component:179; Jaccard:0.25601
 |
Correlation:0.046982
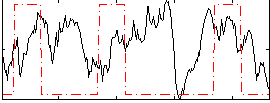  |
Correlation:0.046982
  |
Correlation:0.24395
  |
Correlation:0.24395
  |
Correlation:0.24395
  |
Correlation:0.24395
  |
Correlation:0.24395
  |
Component:179; Jaccard:0.15129
 |
Component:179; Jaccard:0.20451
 |
Component:371; Jaccard:0.25253
 |
Component:371; Jaccard:0.26281
 |
Component:371; Jaccard:0.25015
 |
Component:371; Jaccard:0.2186
 |
Component:395; Jaccard:0.18544
 |
Correlation:0.24395
  |
Correlation:0.24395
  |
Correlation:0.046982
  |
Correlation:0.046982
  |
Correlation:0.046982
  |
Correlation:0.046982
  |
Correlation:0.12498
  |
Component:127; Jaccard:0.14956
 |
Component:127; Jaccard:0.18795
 |
Component:127; Jaccard:0.21999
 |
Component:127; Jaccard:0.22341
 |
Component:395; Jaccard:0.21972
 |
Component:395; Jaccard:0.20694
 |
Component:371; Jaccard:0.18101
 |
Correlation:0.019213
  |
Correlation:0.019213
  |
Correlation:0.019213
  |
Correlation:0.019213
  |
Correlation:0.12498
  |
Correlation:0.12498
  |
Correlation:0.046982
  |
Component:337; Jaccard:0.14468
 |
Component:395; Jaccard:0.17673
 |
Component:395; Jaccard:0.20625
 |
Component:395; Jaccard:0.22109
 |
Component:127; Jaccard:0.20592
 |
Component:127; Jaccard:0.1676
 |
Component:127; Jaccard:0.13328
 |
Correlation:0.038094
  |
Correlation:0.12498
  |
Correlation:0.12498
  |
Correlation:0.12498
  |
Correlation:0.019213
  |
Correlation:0.019213
  |
Correlation:0.019213
  |
Component:066; Jaccard:0.14184
 |
Component:066; Jaccard:0.17408
 |
Component:066; Jaccard:0.19928
 |
Component:066; Jaccard:0.2022
 |
Component:066; Jaccard:0.18474
 |
Component:066; Jaccard:0.15715
 |
Component:066; Jaccard:0.12851
 |
Correlation:0.08603
  |
Correlation:0.08603
  |
Correlation:0.08603
  |
Correlation:0.08603
  |
Correlation:0.08603
  |
Correlation:0.08603
  |
Correlation:0.08603
  |
Component:395; Jaccard:0.14102
 |
Component:337; Jaccard:0.17333
 |
Component:337; Jaccard:0.19348
 |
Component:337; Jaccard:0.19398
 |
Component:337; Jaccard:0.17879
 |
Component:337; Jaccard:0.14873
 |
Component:337; Jaccard:0.11741
 |
Correlation:0.12498
  |
Correlation:0.038094
  |
Correlation:0.038094
  |
Correlation:0.038094
  |
Correlation:0.038094
  |
Correlation:0.038094
  |
Correlation:0.038094
  |
Component:270; Jaccard:0.13309
 |
Component:270; Jaccard:0.15644
 |
Component:270; Jaccard:0.17047
 |
Component:270; Jaccard:0.16606
 |
Component:270; Jaccard:0.14657
 |
Component:270; Jaccard:0.11452
 |
Component:270; Jaccard:0.088435
 |
Correlation:0.011654
  |
Correlation:0.011654
  |
Correlation:0.011654
  |
Correlation:0.011654
  |
Correlation:0.011654
  |
Correlation:0.011654
  |
Correlation:0.011654
  |
Component:255; Jaccard:0.1256
 |
Component:255; Jaccard:0.14483
 |
Component:255; Jaccard:0.15338
 |
Component:255; Jaccard:0.14296
 |
Component:255; Jaccard:0.11674
 |
Component:255; Jaccard:0.087101
 |
Component:302; Jaccard:0.068905
 |
Correlation:-0.098864
  |
Correlation:-0.098864
  |
Correlation:-0.098864
  |
Correlation:-0.098864
  |
Correlation:-0.098864
  |
Correlation:-0.098864
  |
Correlation:0.18035
  |
Component:131; Jaccard:0.10481
 |
Component:217; Jaccard:0.11706
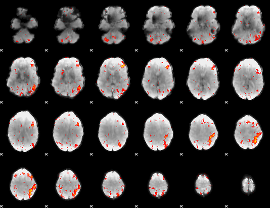 |
Component:217; Jaccard:0.12371
 |
Component:217; Jaccard:0.11738
 |
Component:217; Jaccard:0.099291
 |
Component:302; Jaccard:0.078694
 |
Component:255; Jaccard:0.061255
 |
Correlation:-0.10126
  |
Correlation:-0.0027289
  |
Correlation:-0.0027289
  |
Correlation:-0.0027289
  |
Correlation:-0.0027289
  |
Correlation:0.18035
  |
Correlation:-0.098864
  |
Component:217; Jaccard:0.10214
 |
Component:386; Jaccard:0.11557
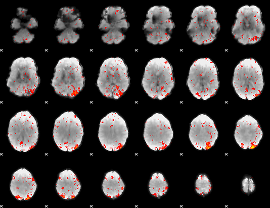 |
Component:386; Jaccard:0.12208
 |
Component:386; Jaccard:0.11415
 |
Component:386; Jaccard:0.096475
 |
Component:217; Jaccard:0.076987
 |
Component:004; Jaccard:0.059624
 |
Correlation:-0.0027289
  |
Correlation:0.071156
  |
Correlation:0.071156
  |
Correlation:0.071156
  |
Correlation:0.071156
  |
Correlation:-0.0027289
  |
Correlation:0.10423
  |
Cope: 03:"MATCH-REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
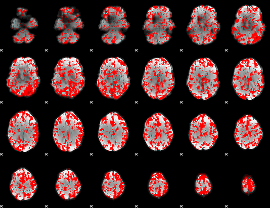 |
GLM binary result thresholded by 2
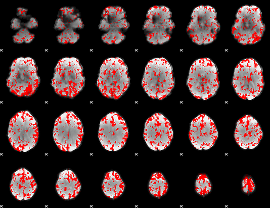 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
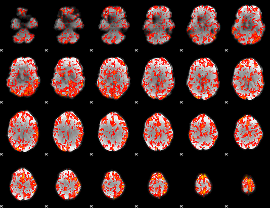 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
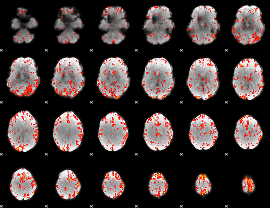 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:197; Jaccard:0.11812
 |
Component:197; Jaccard:0.14976
 |
Component:197; Jaccard:0.18991
 |
Component:197; Jaccard:0.23178
 |
Component:197; Jaccard:0.25022
 |
Component:197; Jaccard:0.22937
 |
Component:197; Jaccard:0.17191
 |
Correlation:0.27746
  |
Correlation:0.27746
  |
Correlation:0.27746
  |
Correlation:0.27746
  |
Correlation:0.27746
  |
Correlation:0.27746
  |
Correlation:0.27746
  |
Component:243; Jaccard:0.09802
 |
Component:243; Jaccard:0.11553
 |
Component:243; Jaccard:0.13373
 |
Component:243; Jaccard:0.14682
 |
Component:243; Jaccard:0.14258
 |
Component:243; Jaccard:0.12293
 |
Component:243; Jaccard:0.093143
 |
Correlation:0.29415
  |
Correlation:0.29415
  |
Correlation:0.29415
  |
Correlation:0.29415
  |
Correlation:0.29415
  |
Correlation:0.29415
  |
Correlation:0.29415
  |
Component:275; Jaccard:0.089456
 |
Component:119; Jaccard:0.098765
 |
Component:119; Jaccard:0.1089
 |
Component:127; Jaccard:0.11987
 |
Component:127; Jaccard:0.12103
 |
Component:127; Jaccard:0.10639
 |
Component:127; Jaccard:0.077546
 |
Correlation:0.21512
  |
Correlation:0.10696
  |
Correlation:0.10696
  |
Correlation:0.19936
  |
Correlation:0.19936
  |
Correlation:0.19936
  |
Correlation:0.19936
  |
Component:119; Jaccard:0.087161
 |
Component:275; Jaccard:0.09856
 |
Component:127; Jaccard:0.10788
 |
Component:119; Jaccard:0.113
 |
Component:119; Jaccard:0.10613
 |
Component:119; Jaccard:0.09077
 |
Component:119; Jaccard:0.064606
 |
Correlation:0.10696
  |
Correlation:0.21512
  |
Correlation:0.19936
  |
Correlation:0.10696
  |
Correlation:0.10696
  |
Correlation:0.10696
  |
Correlation:0.10696
  |
Component:002; Jaccard:0.083576
 |
Component:002; Jaccard:0.094772
 |
Component:275; Jaccard:0.10726
 |
Component:275; Jaccard:0.11171
 |
Component:275; Jaccard:0.10441
 |
Component:002; Jaccard:0.086863
 |
Component:002; Jaccard:0.064035
 |
Correlation:0.33914
  |
Correlation:0.33914
  |
Correlation:0.21512
  |
Correlation:0.21512
  |
Correlation:0.21512
  |
Correlation:0.33914
  |
Correlation:0.33914
  |
Component:305; Jaccard:0.080987
 |
Component:127; Jaccard:0.093046
 |
Component:002; Jaccard:0.10446
 |
Component:002; Jaccard:0.10805
 |
Component:002; Jaccard:0.10233
 |
Component:275; Jaccard:0.086221
 |
Component:275; Jaccard:0.063254
 |
Correlation:0.19193
  |
Correlation:0.19936
  |
Correlation:0.33914
  |
Correlation:0.33914
  |
Correlation:0.33914
  |
Correlation:0.21512
  |
Correlation:0.21512
  |
Component:282; Jaccard:0.080884
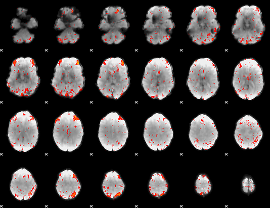 |
Component:282; Jaccard:0.089921
 |
Component:376; Jaccard:0.095981
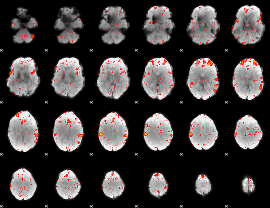 |
Component:376; Jaccard:0.097949
 |
Component:376; Jaccard:0.089615
 |
Component:258; Jaccard:0.075549
 |
Component:258; Jaccard:0.060743
 |
Correlation:0.26164
  |
Correlation:0.26164
  |
Correlation:0.18373
 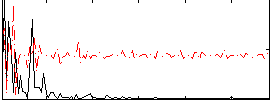 |
Correlation:0.18373
  |
Correlation:0.18373
  |
Correlation:0.22563
  |
Correlation:0.22563
  |
Component:127; Jaccard:0.079675
 |
Component:376; Jaccard:0.088234
 |
Component:282; Jaccard:0.09572
 |
Component:282; Jaccard:0.096381
 |
Component:305; Jaccard:0.088376
 |
Component:376; Jaccard:0.072345
 |
Component:338; Jaccard:0.051978
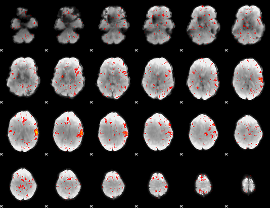 |
Correlation:0.19936
  |
Correlation:0.18373
  |
Correlation:0.26164
  |
Correlation:0.26164
  |
Correlation:0.19193
  |
Correlation:0.18373
  |
Correlation:0.23569
  |
Component:096; Jaccard:0.079321
 |
Component:305; Jaccard:0.088177
 |
Component:305; Jaccard:0.093794
 |
Component:305; Jaccard:0.094765
 |
Component:131; Jaccard:0.085647
 |
Component:338; Jaccard:0.070523
 |
Component:209; Jaccard:0.050443
 |
Correlation:0.25741
  |
Correlation:0.19193
  |
Correlation:0.19193
  |
Correlation:0.19193
  |
Correlation:0.24952
  |
Correlation:0.23569
  |
Correlation:0.2281
  |
Component:376; Jaccard:0.079261
 |
Component:131; Jaccard:0.085204
 |
Component:131; Jaccard:0.091707
 |
Component:131; Jaccard:0.092189
 |
Component:282; Jaccard:0.08476
 |
Component:131; Jaccard:0.069562
 |
Component:096; Jaccard:0.049093
 |
Correlation:0.18373
  |
Correlation:0.24952
  |
Correlation:0.24952
  |
Correlation:0.24952
  |
Correlation:0.26164
  |
Correlation:0.24952
  |
Correlation:0.25741
  |
Cope: 04:"REL-MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
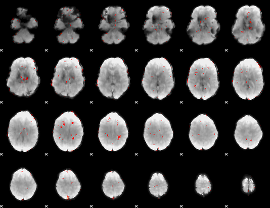 |
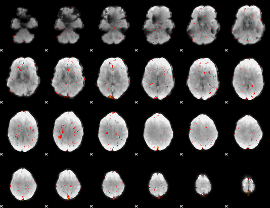
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:046; Jaccard:0.098892
 |
Component:046; Jaccard:0.10631
 |
Component:297; Jaccard:0.097441
 |
Component:297; Jaccard:0.076312
 |
Component:297; Jaccard:0.044319
 |
Component:297; Jaccard:0.02115
 |
Component:277; Jaccard:0.010944
 |
Correlation:0.084238
 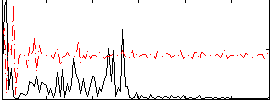 |
Correlation:0.084238
  |
Correlation:0.31733
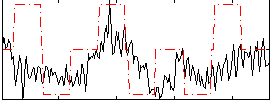 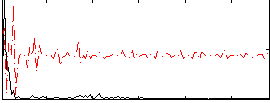 |
Correlation:0.31733
  |
Correlation:0.31733
  |
Correlation:0.31733
  |
Correlation:0.4577
  |
Component:297; Jaccard:0.085021
 |
Component:297; Jaccard:0.099037
 |
Component:046; Jaccard:0.096755
 |
Component:046; Jaccard:0.06728
 |
Component:277; Jaccard:0.037431
 |
Component:277; Jaccard:0.019425
 |
Component:297; Jaccard:0.008986
 |
Correlation:0.31733
  |
Correlation:0.31733
  |
Correlation:0.084238
  |
Correlation:0.084238
  |
Correlation:0.4577
  |
Correlation:0.4577
  |
Correlation:0.31733
  |
Component:211; Jaccard:0.081585
 |
Component:296; Jaccard:0.085884
 |
Component:296; Jaccard:0.07619
 |
Component:277; Jaccard:0.054667
 |
Component:046; Jaccard:0.02902
 |
Component:296; Jaccard:0.013716
 |
Component:296; Jaccard:0.004918
 |
Correlation:0.17468
  |
Correlation:0.14839
  |
Correlation:0.14839
  |
Correlation:0.4577
  |
Correlation:0.084238
  |
Correlation:0.14839
  |
Correlation:0.14839
  |
Component:082; Jaccard:0.080995
 |
Component:211; Jaccard:0.084104
 |
Component:211; Jaccard:0.073733
 |
Component:001; Jaccard:0.053412
 |
Component:001; Jaccard:0.028494
 |
Component:046; Jaccard:0.010742
 |
Component:042; Jaccard:0.00429
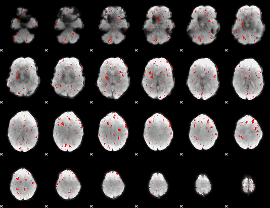 |
Correlation:0.060938
  |
Correlation:0.17468
  |
Correlation:0.17468
  |
Correlation:0.22362
 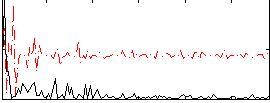 |
Correlation:0.22362
  |
Correlation:0.084238
  |
Correlation:0.077287
 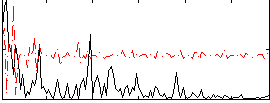 |
Component:169; Jaccard:0.079358
 |
Component:082; Jaccard:0.082017
 |
Component:001; Jaccard:0.072686
 |
Component:211; Jaccard:0.053262
 |
Component:296; Jaccard:0.027718
 |
Component:001; Jaccard:0.01049
 |
Component:306; Jaccard:0.004082
 |
Correlation:0.074143
  |
Correlation:0.060938
  |
Correlation:0.22362
  |
Correlation:0.17468
  |
Correlation:0.14839
  |
Correlation:0.22362
  |
Correlation:0.065196
  |
Component:296; Jaccard:0.079299
 |
Component:001; Jaccard:0.077265
 |
Component:277; Jaccard:0.065614
 |
Component:296; Jaccard:0.052055
 |
Component:211; Jaccard:0.025889
 |
Component:211; Jaccard:0.010065
 |
Component:211; Jaccard:0.003667
 |
Correlation:0.14839
  |
Correlation:0.22362
  |
Correlation:0.4577
  |
Correlation:0.14839
  |
Correlation:0.17468
  |
Correlation:0.17468
  |
Correlation:0.17468
  |
Component:310; Jaccard:0.078065
 |
Component:169; Jaccard:0.074918
 |
Component:082; Jaccard:0.064426
 |
Component:315; Jaccard:0.041858
 |
Component:315; Jaccard:0.017916
 |
Component:136; Jaccard:0.00781
 |
Component:146; Jaccard:0.003536
 |
Correlation:0.12899
  |
Correlation:0.074143
  |
Correlation:0.060938
  |
Correlation:0.14502
  |
Correlation:0.14502
  |
Correlation:0.21916
  |
Correlation:-0.053893
  |
Component:136; Jaccard:0.077122
 |
Component:310; Jaccard:0.073433
 |
Component:315; Jaccard:0.059617
 |
Component:082; Jaccard:0.035978
 |
Component:355; Jaccard:0.016949
 |
Component:355; Jaccard:0.007364
 |
Component:097; Jaccard:0.003445
 |
Correlation:0.21916
  |
Correlation:0.12899
  |
Correlation:0.14502
  |
Correlation:0.060938
  |
Correlation:0.0068865
  |
Correlation:0.0068865
  |
Correlation:-0.029672
  |
Component:015; Jaccard:0.072737
 |
Component:136; Jaccard:0.072138
 |
Component:169; Jaccard:0.058745
 |
Component:015; Jaccard:0.035784
 |
Component:136; Jaccard:0.016935
 |
Component:306; Jaccard:0.007032
 |
Component:001; Jaccard:0.003369
 |
Correlation:0.11352
  |
Correlation:0.21916
  |
Correlation:0.074143
  |
Correlation:0.11352
  |
Correlation:0.21916
  |
Correlation:0.065196
  |
Correlation:0.22362
  |
Component:355; Jaccard:0.071
 |
Component:015; Jaccard:0.069117
 |
Component:136; Jaccard:0.058398
 |
Component:136; Jaccard:0.035252
 |
Component:015; Jaccard:0.016566
 |
Component:060; Jaccard:0.00703
 |
Component:355; Jaccard:0.003163
 |
Correlation:0.0068865
  |
Correlation:0.11352
  |
Correlation:0.21916
  |
Correlation:0.21916
  |
Correlation:0.11352
  |
Correlation:0.10716
  |
Correlation:0.0068865
  |
Cope: 05:"neg_MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
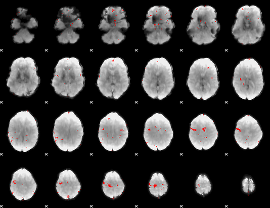 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
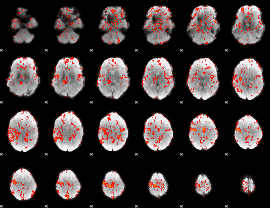 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
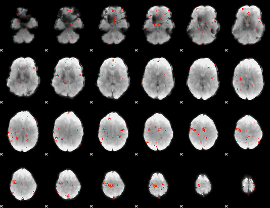 |
GLM Z-score result thresholded by 3.5
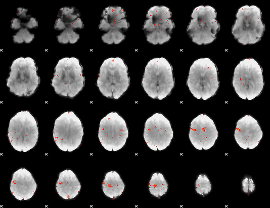 |
GLM Z-score result thresholded by 4
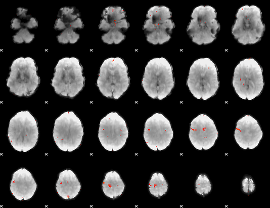 |
Component:216; Jaccard:0.092902
 |
Component:216; Jaccard:0.094251
 |
Component:368; Jaccard:0.092501
 |
Component:315; Jaccard:0.084445
 |
Component:368; Jaccard:0.07033
 |
Component:368; Jaccard:0.05083
 |
Component:368; Jaccard:0.031241
 |
Correlation:0.027414
  |
Correlation:0.027414
  |
Correlation:-0.054197
  |
Correlation:0.24743
  |
Correlation:-0.054197
  |
Correlation:-0.054197
  |
Correlation:-0.054197
  |
Component:367; Jaccard:0.086026
 |
Component:368; Jaccard:0.092022
 |
Component:082; Jaccard:0.090157
 |
Component:368; Jaccard:0.084393
 |
Component:315; Jaccard:0.067774
 |
Component:315; Jaccard:0.044743
 |
Component:162; Jaccard:0.025118
 |
Correlation:0.077543
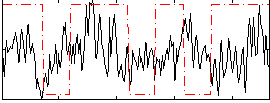 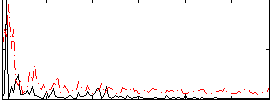 |
Correlation:-0.054197
  |
Correlation:0.097685
 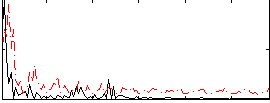 |
Correlation:-0.054197
  |
Correlation:0.24743
  |
Correlation:0.24743
  |
Correlation:0.0071131
  |
Component:368; Jaccard:0.084615
 |
Component:367; Jaccard:0.091393
 |
Component:367; Jaccard:0.088934
 |
Component:082; Jaccard:0.08012
 |
Component:082; Jaccard:0.058389
 |
Component:162; Jaccard:0.041684
 |
Component:314; Jaccard:0.024815
 |
Correlation:-0.054197
  |
Correlation:0.077543
  |
Correlation:0.077543
  |
Correlation:0.097685
  |
Correlation:0.097685
  |
Correlation:0.0071131
  |
Correlation:0.15661
  |
Component:314; Jaccard:0.078854
 |
Component:082; Jaccard:0.08645
 |
Component:315; Jaccard:0.08734
 |
Component:367; Jaccard:0.075774
 |
Component:314; Jaccard:0.05584
 |
Component:314; Jaccard:0.038183
 |
Component:315; Jaccard:0.023512
 |
Correlation:0.15661
  |
Correlation:0.097685
  |
Correlation:0.24743
  |
Correlation:0.077543
  |
Correlation:0.15661
  |
Correlation:0.15661
  |
Correlation:0.24743
  |
Component:082; Jaccard:0.076307
 |
Component:314; Jaccard:0.084225
 |
Component:216; Jaccard:0.085334
 |
Component:314; Jaccard:0.075696
 |
Component:297; Jaccard:0.055312
 |
Component:216; Jaccard:0.037186
 |
Component:216; Jaccard:0.023441
 |
Correlation:0.097685
  |
Correlation:0.15661
  |
Correlation:0.027414
  |
Correlation:0.15661
  |
Correlation:0.27552
  |
Correlation:0.027414
  |
Correlation:0.027414
  |
Component:307; Jaccard:0.074138
 |
Component:307; Jaccard:0.080341
 |
Component:314; Jaccard:0.084274
 |
Component:307; Jaccard:0.074393
 |
Component:367; Jaccard:0.055297
 |
Component:297; Jaccard:0.037088
 |
Component:082; Jaccard:0.021461
 |
Correlation:0.070907
  |
Correlation:0.070907
  |
Correlation:0.15661
  |
Correlation:0.070907
  |
Correlation:0.077543
  |
Correlation:0.27552
  |
Correlation:0.097685
  |
Component:275; Jaccard:0.070806
 |
Component:315; Jaccard:0.079631
 |
Component:307; Jaccard:0.082031
 |
Component:216; Jaccard:0.071686
 |
Component:307; Jaccard:0.055018
 |
Component:082; Jaccard:0.035492
 |
Component:297; Jaccard:0.020539
 |
Correlation:-0.017157
  |
Correlation:0.24743
  |
Correlation:0.070907
  |
Correlation:0.027414
  |
Correlation:0.070907
  |
Correlation:0.097685
  |
Correlation:0.27552
  |
Component:153; Jaccard:0.069612
 |
Component:092; Jaccard:0.073731
 |
Component:092; Jaccard:0.073524
 |
Component:092; Jaccard:0.068762
 |
Component:216; Jaccard:0.054769
 |
Component:367; Jaccard:0.034439
 |
Component:277; Jaccard:0.018502
 |
Correlation:0.075472
  |
Correlation:0.14927
  |
Correlation:0.14927
  |
Correlation:0.14927
  |
Correlation:0.027414
  |
Correlation:0.077543
  |
Correlation:0.39845
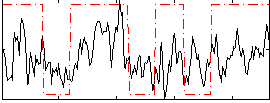 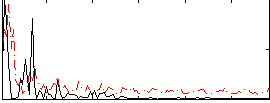 |
Component:315; Jaccard:0.069496
 |
Component:162; Jaccard:0.068378
 |
Component:297; Jaccard:0.071316
 |
Component:297; Jaccard:0.068706
 |
Component:162; Jaccard:0.054402
 |
Component:307; Jaccard:0.033079
 |
Component:367; Jaccard:0.018268
 |
Correlation:0.24743
  |
Correlation:0.0071131
  |
Correlation:0.27552
  |
Correlation:0.27552
  |
Correlation:0.0071131
  |
Correlation:0.070907
  |
Correlation:0.077543
  |
Component:092; Jaccard:0.067731
 |
Component:297; Jaccard:0.06731
 |
Component:162; Jaccard:0.068571
 |
Component:162; Jaccard:0.064617
 |
Component:092; Jaccard:0.052576
 |
Component:277; Jaccard:0.032564
 |
Component:092; Jaccard:0.016098
 |
Correlation:0.14927
  |
Correlation:0.27552
  |
Correlation:0.0071131
  |
Correlation:0.0071131
  |
Correlation:0.14927
  |
Correlation:0.39845
  |
Correlation:0.14927
  |
Cope: 06:"neg_REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
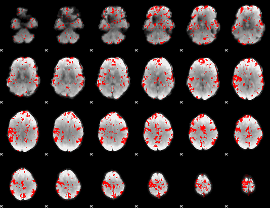 |
GLM binary result thresholded by 2.5
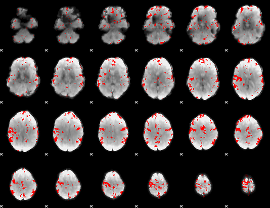 |
GLM binary result thresholded by 3
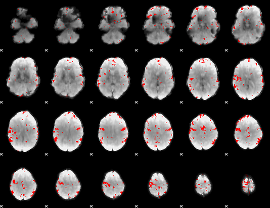 |
GLM binary result thresholded by 3.5
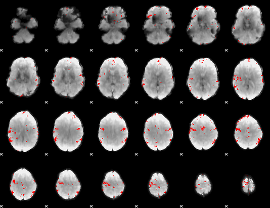 |
GLM binary result thresholded by 4
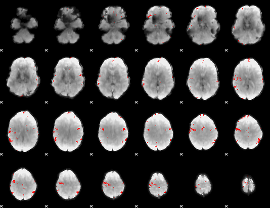 |
GLM Z-score result thresholded by 1
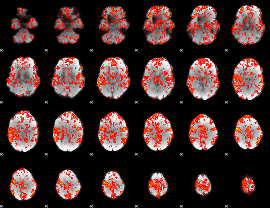 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:275; Jaccard:0.1386
 |
Component:275; Jaccard:0.16499
 |
Component:275; Jaccard:0.18259
 |
Component:275; Jaccard:0.18159
 |
Component:275; Jaccard:0.15665
 |
Component:275; Jaccard:0.1176
 |
Component:275; Jaccard:0.078198
 |
Correlation:0.34579
  |
Correlation:0.34579
  |
Correlation:0.34579
  |
Correlation:0.34579
  |
Correlation:0.34579
  |
Correlation:0.34579
  |
Correlation:0.34579
  |
Component:216; Jaccard:0.11202
 |
Component:216; Jaccard:0.13323
 |
Component:216; Jaccard:0.14891
 |
Component:216; Jaccard:0.15385
 |
Component:216; Jaccard:0.13905
 |
Component:216; Jaccard:0.10788
 |
Component:216; Jaccard:0.074293
 |
Correlation:0.25606
  |
Correlation:0.25606
  |
Correlation:0.25606
  |
Correlation:0.25606
  |
Correlation:0.25606
  |
Correlation:0.25606
  |
Correlation:0.25606
  |
Component:324; Jaccard:0.092273
 |
Component:368; Jaccard:0.10718
 |
Component:368; Jaccard:0.12361
 |
Component:368; Jaccard:0.12988
 |
Component:368; Jaccard:0.12143
 |
Component:368; Jaccard:0.097746
 |
Component:368; Jaccard:0.069333
 |
Correlation:0.26527
  |
Correlation:0.18536
  |
Correlation:0.18536
  |
Correlation:0.18536
  |
Correlation:0.18536
  |
Correlation:0.18536
  |
Correlation:0.18536
  |
Component:368; Jaccard:0.087555
 |
Component:324; Jaccard:0.10369
 |
Component:324; Jaccard:0.11109
 |
Component:324; Jaccard:0.10561
 |
Component:051; Jaccard:0.090917
 |
Component:051; Jaccard:0.07625
 |
Component:051; Jaccard:0.056514
 |
Correlation:0.18536
  |
Correlation:0.26527
  |
Correlation:0.26527
  |
Correlation:0.26527
  |
Correlation:0.3405
  |
Correlation:0.3405
  |
Correlation:0.3405
  |
Component:103; Jaccard:0.085892
 |
Component:143; Jaccard:0.096951
 |
Component:143; Jaccard:0.10522
 |
Component:143; Jaccard:0.10126
 |
Component:324; Jaccard:0.089169
 |
Component:324; Jaccard:0.067284
 |
Component:324; Jaccard:0.044662
 |
Correlation:0.20524
  |
Correlation:0.18446
  |
Correlation:0.18446
  |
Correlation:0.18446
  |
Correlation:0.26527
  |
Correlation:0.26527
  |
Correlation:0.26527
  |
Component:190; Jaccard:0.085411
 |
Component:190; Jaccard:0.095387
 |
Component:190; Jaccard:0.0995
 |
Component:051; Jaccard:0.09743
 |
Component:143; Jaccard:0.083333
 |
Component:092; Jaccard:0.062626
 |
Component:092; Jaccard:0.042155
 |
Correlation:0.073153
  |
Correlation:0.073153
  |
Correlation:0.073153
  |
Correlation:0.3405
  |
Correlation:0.18446
  |
Correlation:0.11036
  |
Correlation:0.11036
  |
Component:143; Jaccard:0.084817
 |
Component:367; Jaccard:0.093198
 |
Component:248; Jaccard:0.098374
 |
Component:367; Jaccard:0.093394
 |
Component:367; Jaccard:0.079417
 |
Component:367; Jaccard:0.060491
 |
Component:155; Jaccard:0.04192
 |
Correlation:0.18446
  |
Correlation:0.10227
  |
Correlation:0.25492
  |
Correlation:0.10227
  |
Correlation:0.10227
  |
Correlation:0.10227
  |
Correlation:0.20949
  |
Component:308; Jaccard:0.08476
 |
Component:308; Jaccard:0.092582
 |
Component:367; Jaccard:0.096824
 |
Component:190; Jaccard:0.092845
 |
Component:248; Jaccard:0.079028
 |
Component:143; Jaccard:0.060237
 |
Component:367; Jaccard:0.040803
 |
Correlation:0.10634
  |
Correlation:0.10634
  |
Correlation:0.10227
  |
Correlation:0.073153
  |
Correlation:0.25492
  |
Correlation:0.18446
  |
Correlation:0.10227
  |
Component:367; Jaccard:0.082613
 |
Component:103; Jaccard:0.092294
 |
Component:308; Jaccard:0.096795
 |
Component:248; Jaccard:0.092652
 |
Component:092; Jaccard:0.077751
 |
Component:047; Jaccard:0.059564
 |
Component:308; Jaccard:0.038405
 |
Correlation:0.10227
  |
Correlation:0.20524
  |
Correlation:0.10634
  |
Correlation:0.25492
  |
Correlation:0.11036
  |
Correlation:0.26782
  |
Correlation:0.10634
  |
Component:322; Jaccard:0.080062
 |
Component:248; Jaccard:0.090603
 |
Component:103; Jaccard:0.094218
 |
Component:308; Jaccard:0.090637
 |
Component:190; Jaccard:0.076583
 |
Component:155; Jaccard:0.058099
 |
Component:047; Jaccard:0.038333
 |
Correlation:0.46545
  |
Correlation:0.25492
  |
Correlation:0.20524
  |
Correlation:0.10634
  |
Correlation:0.073153
  |
Correlation:0.20949
  |
Correlation:0.26782
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































