Task name: GAMBLING, TR=0.72, totally 253 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"PUNISH" BACK
|
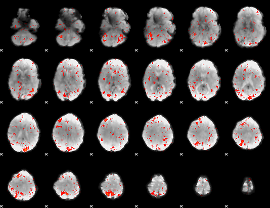
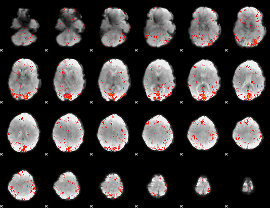
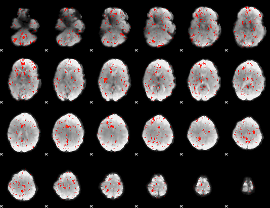
GLM result group-wise
 |
GLM binary result thresholded by 1
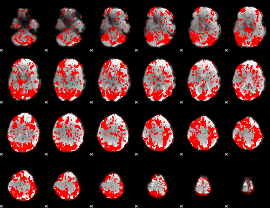 |
GLM binary result thresholded by 1.5
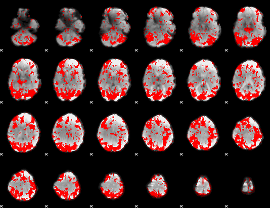 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
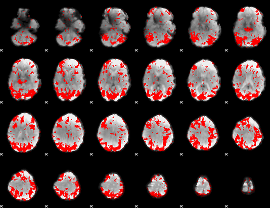
GLM binary result thresholded by 3
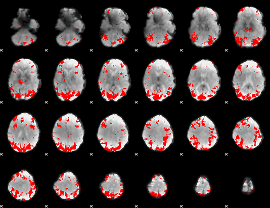 |
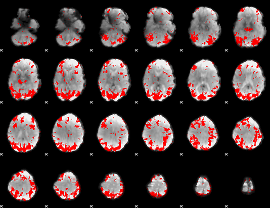
GLM binary result thresholded by 3.5
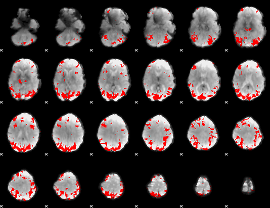 |
GLM binary result thresholded by 4
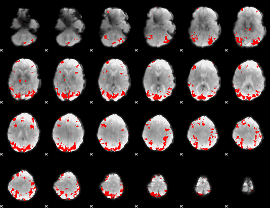 |
GLM Z-score result thresholded by 1
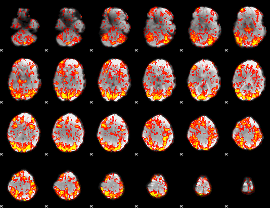 |
GLM Z-score result thresholded by 1.5
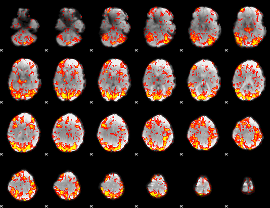 |
GLM Z-score result thresholded by 2
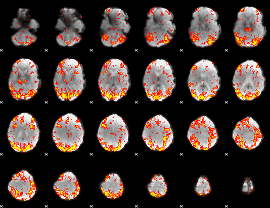 |
GLM Z-score result thresholded by 2.5
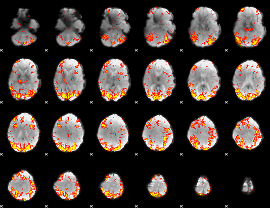 |
GLM Z-score result thresholded by 3
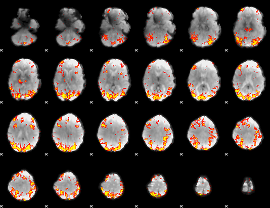 |
GLM Z-score result thresholded by 3.5
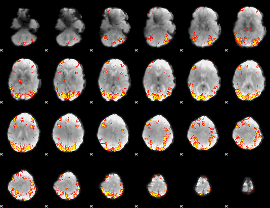 |
GLM Z-score result thresholded by 4
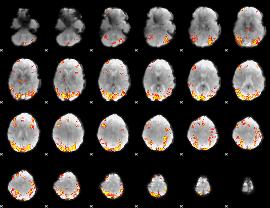 |
Component:395; Jaccard:0.11996
 |
Component:228; Jaccard:0.14619
 |
Component:228; Jaccard:0.18077
 |
Component:228; Jaccard:0.21718
 |
Component:228; Jaccard:0.25147
 |
Component:228; Jaccard:0.2792
 |
Component:228; Jaccard:0.29996
 |
Correlation:0.23259
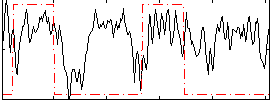 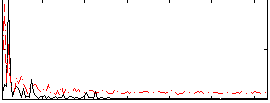 |
Correlation:0.29775
  |
Correlation:0.29775
  |
Correlation:0.29775
  |
Correlation:0.29775
  |
Correlation:0.29775
  |
Correlation:0.29775
  |
Component:228; Jaccard:0.11741
 |
Component:395; Jaccard:0.14051
 |
Component:395; Jaccard:0.16329
 |
Component:259; Jaccard:0.18656
 |
Component:259; Jaccard:0.2219
 |
Component:259; Jaccard:0.25181
 |
Component:259; Jaccard:0.27577
 |
Correlation:0.29775
  |
Correlation:0.23259
  |
Correlation:0.23259
  |
Correlation:0.296
  |
Correlation:0.296
  |
Correlation:0.296
  |
Correlation:0.296
  |
Component:242; Jaccard:0.11149
 |
Component:242; Jaccard:0.12974
 |
Component:259; Jaccard:0.15253
 |
Component:395; Jaccard:0.1822
 |
Component:066; Jaccard:0.20962
 |
Component:066; Jaccard:0.24185
 |
Component:066; Jaccard:0.26887
 |
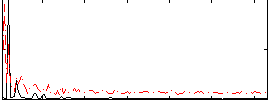
Correlation:0.16873
 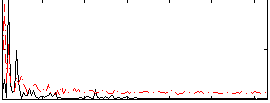 |
Correlation:0.16873
  |
Correlation:0.296
  |
Correlation:0.23259
  |
Correlation:0.35044
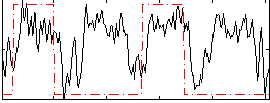  |
Correlation:0.35044
  |
Correlation:0.35044
  |
Component:104; Jaccard:0.10311
 |
Component:259; Jaccard:0.12262
 |
Component:242; Jaccard:0.14968
 |
Component:066; Jaccard:0.17529
 |
Component:395; Jaccard:0.19501
 |
Component:395; Jaccard:0.19804
 |
Component:395; Jaccard:0.19222
 |
Correlation:0.2229
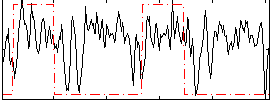  |
Correlation:0.296
  |
Correlation:0.16873
  |
Correlation:0.35044
  |
Correlation:0.23259
  |
Correlation:0.23259
  |
Correlation:0.23259
  |
Component:156; Jaccard:0.099827
 |
Component:104; Jaccard:0.11896
 |
Component:066; Jaccard:0.14224
 |
Component:242; Jaccard:0.16658
 |
Component:242; Jaccard:0.17875
 |
Component:242; Jaccard:0.18324
 |
Component:242; Jaccard:0.17956
 |
Correlation:0.13612
  |
Correlation:0.2229
  |
Correlation:0.35044
  |
Correlation:0.16873
  |
Correlation:0.16873
  |
Correlation:0.16873
  |
Correlation:0.16873
  |
Component:259; Jaccard:0.098207
 |
Component:175; Jaccard:0.11622
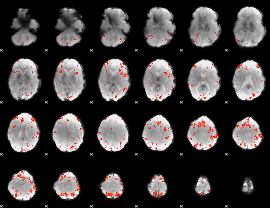 |
Component:104; Jaccard:0.1371
 |
Component:104; Jaccard:0.15381
 |
Component:104; Jaccard:0.16576
 |
Component:104; Jaccard:0.17089
 |
Component:104; Jaccard:0.17095
 |
Correlation:0.296
  |
Correlation:0.19737
  |
Correlation:0.2229
  |
Correlation:0.2229
  |
Correlation:0.2229
  |
Correlation:0.2229
  |
Correlation:0.2229
  |
Component:175; Jaccard:0.097464
 |
Component:066; Jaccard:0.11351
 |
Component:175; Jaccard:0.13563
 |
Component:175; Jaccard:0.15147
 |
Component:175; Jaccard:0.163
 |
Component:175; Jaccard:0.1616
 |
Component:012; Jaccard:0.15516
 |
Correlation:0.19737
  |
Correlation:0.35044
  |
Correlation:0.19737
  |
Correlation:0.19737
  |
Correlation:0.19737
  |
Correlation:0.19737
  |
Correlation:0.15674
 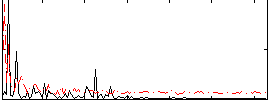 |
Component:231; Jaccard:0.096109
 |
Component:156; Jaccard:0.11207
 |
Component:012; Jaccard:0.12682
 |
Component:012; Jaccard:0.14138
 |
Component:012; Jaccard:0.15244
 |
Component:012; Jaccard:0.15562
 |
Component:175; Jaccard:0.1551
 |
Correlation:0.26982
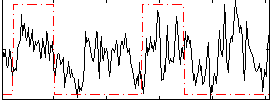 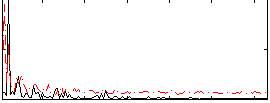 |
Correlation:0.13612
  |
Correlation:0.15674
  |
Correlation:0.15674
  |
Correlation:0.15674
  |
Correlation:0.15674
  |
Correlation:0.19737
  |
Component:012; Jaccard:0.095306
 |
Component:012; Jaccard:0.11099
 |
Component:231; Jaccard:0.12434
 |
Component:237; Jaccard:0.13853
 |
Component:237; Jaccard:0.14768
 |
Component:237; Jaccard:0.15039
 |
Component:251; Jaccard:0.14677
 |
Correlation:0.15674
  |
Correlation:0.15674
  |
Correlation:0.26982
  |
Correlation:0.013792
  |
Correlation:0.013792
  |
Correlation:0.013792
  |
Correlation:0.13212
  |
Component:237; Jaccard:0.094875
 |
Component:231; Jaccard:0.11021
 |
Component:237; Jaccard:0.12428
 |
Component:251; Jaccard:0.13603
 |
Component:251; Jaccard:0.1464
 |
Component:251; Jaccard:0.14943
 |
Component:237; Jaccard:0.14653
 |
Correlation:0.013792
  |
Correlation:0.26982
  |
Correlation:0.013792
  |
Correlation:0.13212
  |
Correlation:0.13212
  |
Correlation:0.13212
  |
Correlation:0.013792
  |
Cope: 02:"REWARD" BACK
|
GLM result group-wise
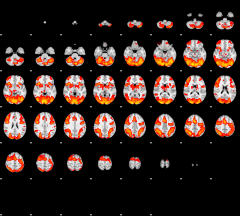 |
GLM binary result thresholded by 1
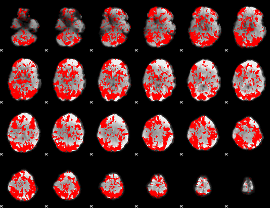 |
GLM binary result thresholded by 1.5
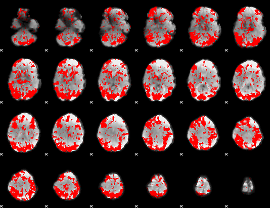 |
GLM binary result thresholded by 2
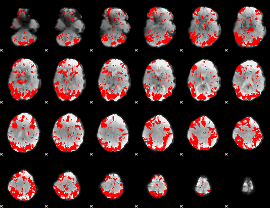 |
GLM binary result thresholded by 2.5
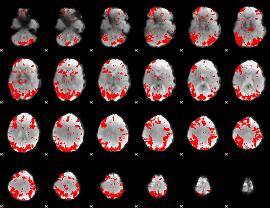 |
GLM binary result thresholded by 3
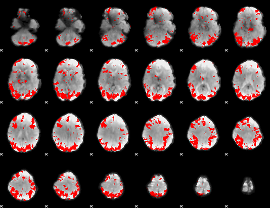 |
GLM binary result thresholded by 3.5
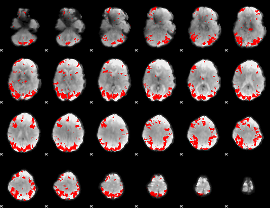 |
GLM binary result thresholded by 4
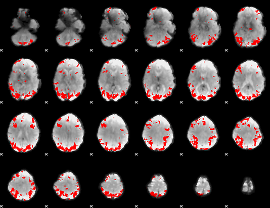 |
GLM Z-score result thresholded by 1
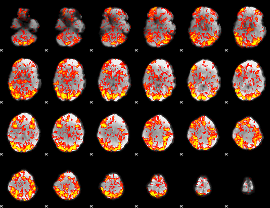 |
GLM Z-score result thresholded by 1.5
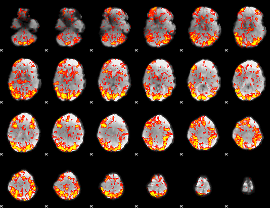 |
GLM Z-score result thresholded by 2
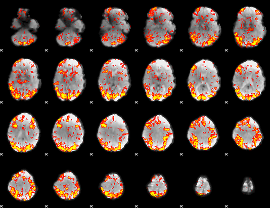 |
GLM Z-score result thresholded by 2.5
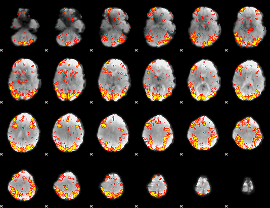 |
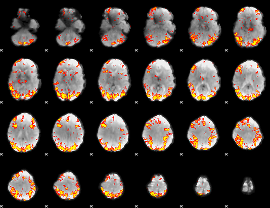
GLM Z-score result thresholded by 3
 |
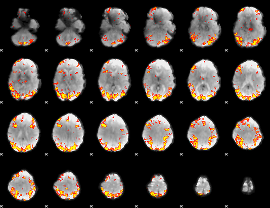
GLM Z-score result thresholded by 3.5
 |
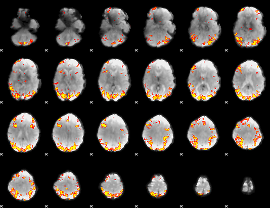
GLM Z-score result thresholded by 4
 |
Component:395; Jaccard:0.1289
 |
Component:242; Jaccard:0.15267
 |
Component:242; Jaccard:0.17638
 |
Component:242; Jaccard:0.19623
 |
Component:242; Jaccard:0.21004
 |
Component:066; Jaccard:0.22688
 |
Component:066; Jaccard:0.24967
 |
Correlation:0.034534
  |
Correlation:0.22036
  |
Correlation:0.22036
  |
Correlation:0.22036
  |
Correlation:0.22036
  |
Correlation:0.27526
  |
Correlation:0.27526
  |
Component:242; Jaccard:0.1283
 |
Component:395; Jaccard:0.15175
 |
Component:395; Jaccard:0.17318
 |
Component:395; Jaccard:0.18979
 |
Component:228; Jaccard:0.20824
 |
Component:259; Jaccard:0.22469
 |
Component:259; Jaccard:0.24537
 |
Correlation:0.22036
  |
Correlation:0.034534
  |
Correlation:0.034534
  |
Correlation:0.034534
  |
Correlation:0.066589
  |
Correlation:0.25057
  |
Correlation:0.25057
  |
Component:325; Jaccard:0.11577
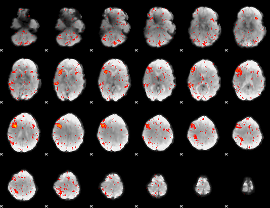 |
Component:228; Jaccard:0.13868
 |
Component:228; Jaccard:0.16329
 |
Component:228; Jaccard:0.18764
 |
Component:395; Jaccard:0.20159
 |
Component:228; Jaccard:0.22461
 |
Component:228; Jaccard:0.23489
 |
Correlation:0.25041
  |
Correlation:0.066589
  |
Correlation:0.066589
  |
Correlation:0.066589
  |
Correlation:0.034534
  |
Correlation:0.066589
  |
Correlation:0.066589
  |
Component:228; Jaccard:0.11497
 |
Component:325; Jaccard:0.13464
 |
Component:012; Jaccard:0.15383
 |
Component:259; Jaccard:0.17475
 |
Component:066; Jaccard:0.20065
 |
Component:242; Jaccard:0.21873
 |
Component:242; Jaccard:0.22061
 |
Correlation:0.066589
  |
Correlation:0.25041
  |
Correlation:0.19779
 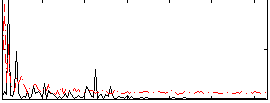 |
Correlation:0.25057
  |
Correlation:0.27526
  |
Correlation:0.22036
  |
Correlation:0.22036
  |
Component:156; Jaccard:0.11334
 |
Component:156; Jaccard:0.13334
 |
Component:156; Jaccard:0.15008
 |
Component:066; Jaccard:0.17158
 |
Component:259; Jaccard:0.20006
 |
Component:395; Jaccard:0.20606
 |
Component:395; Jaccard:0.20795
 |
Correlation:0.10335
 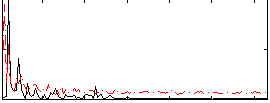 |
Correlation:0.10335
  |
Correlation:0.10335
  |
Correlation:0.27526
  |
Correlation:0.25057
  |
Correlation:0.034534
  |
Correlation:0.034534
  |
Component:012; Jaccard:0.11325
 |
Component:012; Jaccard:0.13315
 |
Component:259; Jaccard:0.14916
 |
Component:012; Jaccard:0.17013
 |
Component:012; Jaccard:0.18123
 |
Component:012; Jaccard:0.18942
 |
Component:012; Jaccard:0.19037
 |
Correlation:0.19779
  |
Correlation:0.19779
  |
Correlation:0.25057
  |
Correlation:0.19779
  |
Correlation:0.19779
  |
Correlation:0.19779
  |
Correlation:0.19779
  |
Component:104; Jaccard:0.10468
 |
Component:259; Jaccard:0.12366
 |
Component:325; Jaccard:0.1484
 |
Component:156; Jaccard:0.16263
 |
Component:251; Jaccard:0.17357
 |
Component:251; Jaccard:0.18259
 |
Component:251; Jaccard:0.18558
 |
Correlation:0.12072
  |
Correlation:0.25057
  |
Correlation:0.25041
  |
Correlation:0.10335
  |
Correlation:0.20973
  |
Correlation:0.20973
  |
Correlation:0.20973
  |
Component:251; Jaccard:0.10145
 |
Component:251; Jaccard:0.12227
 |
Component:066; Jaccard:0.14319
 |
Component:251; Jaccard:0.16032
 |
Component:156; Jaccard:0.16545
 |
Component:132; Jaccard:0.16876
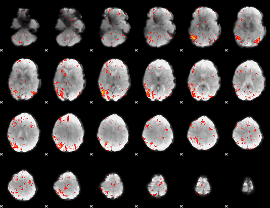 |
Component:132; Jaccard:0.17016
 |
Correlation:0.20973
  |
Correlation:0.20973
  |
Correlation:0.27526
  |
Correlation:0.20973
  |
Correlation:0.10335
  |
Correlation:0.18722
 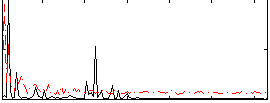 |
Correlation:0.18722
  |
Component:237; Jaccard:0.10143
 |
Component:104; Jaccard:0.12011
 |
Component:251; Jaccard:0.14232
 |
Component:325; Jaccard:0.15763
 |
Component:325; Jaccard:0.16182
 |
Component:104; Jaccard:0.16389
 |
Component:104; Jaccard:0.16697
 |
Correlation:0.2374
  |
Correlation:0.12072
  |
Correlation:0.20973
  |
Correlation:0.25041
  |
Correlation:0.25041
  |
Correlation:0.12072
  |
Correlation:0.12072
  |
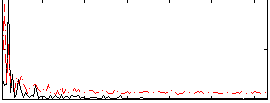
Component:347; Jaccard:0.10008
 |
Component:237; Jaccard:0.11738
 |
Component:104; Jaccard:0.13569
 |
Component:132; Jaccard:0.14939
 |
Component:132; Jaccard:0.16097
 |
Component:156; Jaccard:0.16128
 |
Component:237; Jaccard:0.15668
 |
Correlation:0.033371
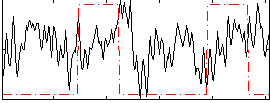 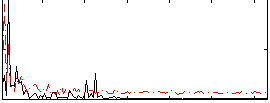 |
Correlation:0.2374
  |
Correlation:0.12072
  |
Correlation:0.18722
  |
Correlation:0.18722
  |
Correlation:0.10335
  |
Correlation:0.2374
  |
Cope: 03:"PUNISH-REWARD" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
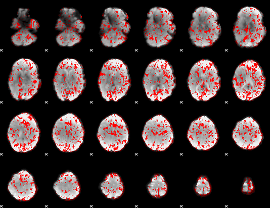 |
GLM binary result thresholded by 1.5
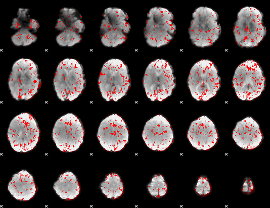 |
GLM binary result thresholded by 2
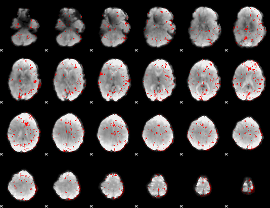 |
GLM binary result thresholded by 2.5
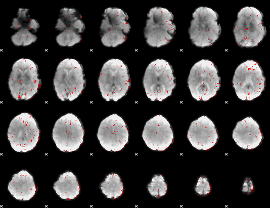 |
GLM binary result thresholded by 3
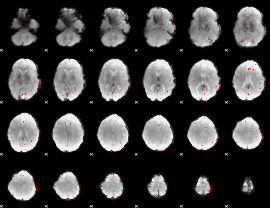 |
GLM binary result thresholded by 3.5
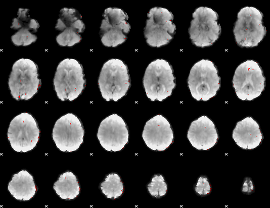 |
GLM binary result thresholded by 4
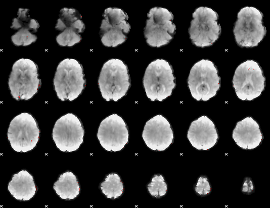 |
GLM Z-score result thresholded by 1
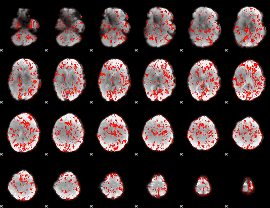 |
GLM Z-score result thresholded by 1.5
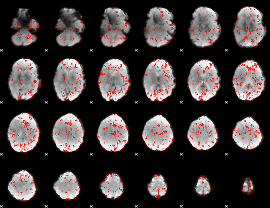 |
GLM Z-score result thresholded by 2
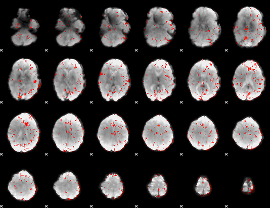 |
GLM Z-score result thresholded by 2.5
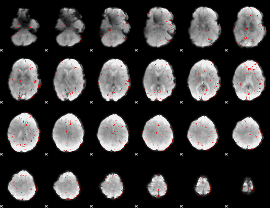 |
GLM Z-score result thresholded by 3
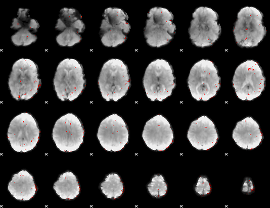 |
GLM Z-score result thresholded by 3.5
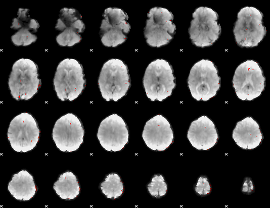 |
GLM Z-score result thresholded by 4
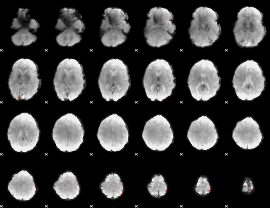 |
Component:070; Jaccard:0.11554
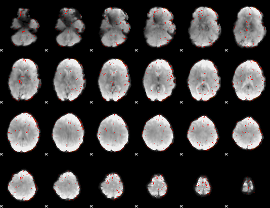 |
Component:378; Jaccard:0.1493
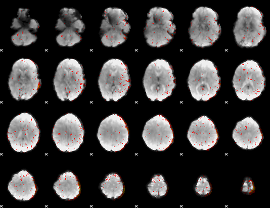 |
Component:378; Jaccard:0.17683
 |
Component:378; Jaccard:0.17382
 |
Component:378; Jaccard:0.12514
 |
Component:378; Jaccard:0.062873
 |
Component:378; Jaccard:0.025774
 |
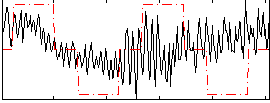
Correlation:0.13931
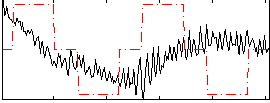 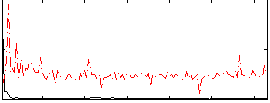 |
Correlation:0.18177
  |
Correlation:0.18177
  |
Correlation:0.18177
  |
Correlation:0.18177
  |
Correlation:0.18177
  |
Correlation:0.18177
  |
Component:378; Jaccard:0.11335
 |
Component:070; Jaccard:0.13671
 |
Component:070; Jaccard:0.14268
 |
Component:070; Jaccard:0.12609
 |
Component:070; Jaccard:0.082436
 |
Component:070; Jaccard:0.040436
 |
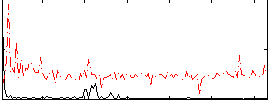
Component:222; Jaccard:0.016351
 |
Correlation:0.18177
  |
Correlation:0.13931
  |
Correlation:0.13931
  |
Correlation:0.13931
  |
Correlation:0.13931
  |
Correlation:0.13931
  |
Correlation:0.064911
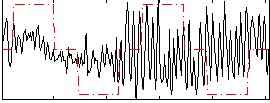 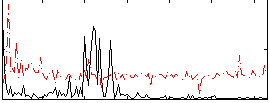 |
Component:116; Jaccard:0.10107
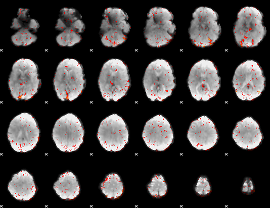 |
Component:116; Jaccard:0.11607
 |
Component:116; Jaccard:0.1212
 |
Component:116; Jaccard:0.10055
 |
Component:222; Jaccard:0.070602
 |
Component:222; Jaccard:0.037133
 |
Component:070; Jaccard:0.015186
 |
Correlation:0.0016139
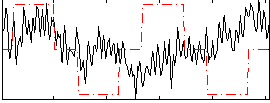 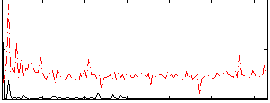 |
Correlation:0.0016139
  |
Correlation:0.0016139
  |
Correlation:0.0016139
  |
Correlation:0.064911
  |
Correlation:0.064911
  |
Correlation:0.13931
  |
Component:199; Jaccard:0.087278
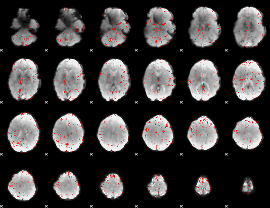 |
Component:264; Jaccard:0.094025
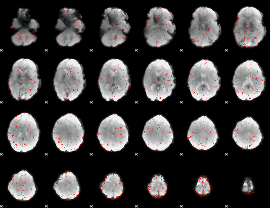 |
Component:264; Jaccard:0.1003
 |
Component:222; Jaccard:0.091173
 |
Component:116; Jaccard:0.067165
 |
Component:116; Jaccard:0.033781
 |
Component:116; Jaccard:0.013388
 |
Correlation:0.23053
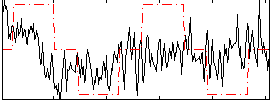 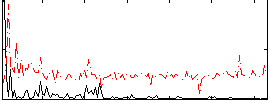 |
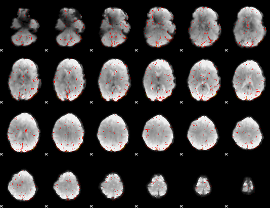
Correlation:0.28155
  |
Correlation:0.28155
  |
Correlation:0.064911
  |
Correlation:0.0016139
  |
Correlation:0.0016139
  |
Correlation:0.0016139
  |
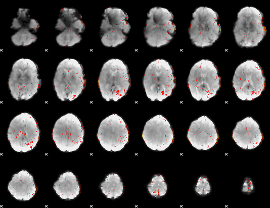
Component:248; Jaccard:0.084055
 |
Component:199; Jaccard:0.092589
 |
Component:222; Jaccard:0.09852
 |
Component:264; Jaccard:0.086977
 |
Component:264; Jaccard:0.058426
 |
Component:191; Jaccard:0.027957
 |
Component:191; Jaccard:0.011781
 |
Correlation:0.13601
  |
Correlation:0.23053
  |
Correlation:0.064911
  |
Correlation:0.28155
  |
Correlation:0.28155
  |
Correlation:-0.02212
  |
Correlation:-0.02212
  |
Component:264; Jaccard:0.079373
 |
Component:248; Jaccard:0.092388
 |
Component:248; Jaccard:0.091594
 |
Component:327; Jaccard:0.076889
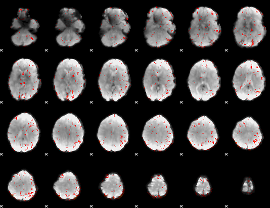 |
Component:327; Jaccard:0.051939
 |
Component:264; Jaccard:0.027322
 |
Component:005; Jaccard:0.011613
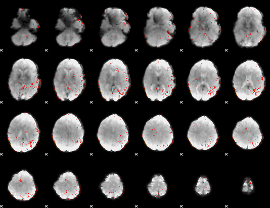 |
Correlation:0.28155
  |
Correlation:0.13601
  |
Correlation:0.13601
  |
Correlation:0.25286
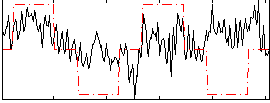 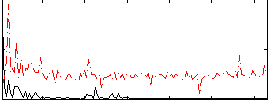 |
Correlation:0.25286
  |
Correlation:0.28155
  |
Correlation:0.13412
  |
Component:327; Jaccard:0.07765
 |
Component:222; Jaccard:0.0878
 |
Component:327; Jaccard:0.087825
 |
Component:393; Jaccard:0.07536
 |
Component:393; Jaccard:0.04984
 |
Component:005; Jaccard:0.027076
 |
Component:327; Jaccard:0.009717
 |
Correlation:0.25286
  |
Correlation:0.064911
  |
Correlation:0.25286
  |
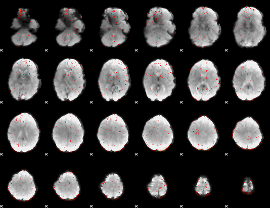
Correlation:0.093189
  |
Correlation:0.093189
  |
Correlation:0.13412
  |
Correlation:0.25286
  |
Component:222; Jaccard:0.074811
 |
Component:327; Jaccard:0.0863
 |
Component:199; Jaccard:0.084155
 |
Component:248; Jaccard:0.072267
 |
Component:005; Jaccard:0.04955
 |
Component:327; Jaccard:0.024712
 |
Component:264; Jaccard:0.009486
 |
Correlation:0.064911
  |
Correlation:0.25286
  |
Correlation:0.23053
  |
Correlation:0.13601
  |
Correlation:0.13412
  |
Correlation:0.25286
  |
Correlation:0.28155
  |
Component:002; Jaccard:0.071894
 |
Component:393; Jaccard:0.076582
 |
Component:393; Jaccard:0.08162
 |
Component:005; Jaccard:0.071234
 |
Component:191; Jaccard:0.046327
 |
Component:393; Jaccard:0.021439
 |
Component:199; Jaccard:0.007705
 |
Correlation:0.17937
  |
Correlation:0.093189
  |
Correlation:0.093189
  |
Correlation:0.13412
  |
Correlation:-0.02212
  |
Correlation:0.093189
  |
Correlation:0.23053
  |
Component:005; Jaccard:0.06605
 |
Component:005; Jaccard:0.076083
 |
Component:005; Jaccard:0.081327
 |
Component:199; Jaccard:0.063327
 |
Component:248; Jaccard:0.040338
 |
Component:199; Jaccard:0.018925
 |
Component:031; Jaccard:0.007621
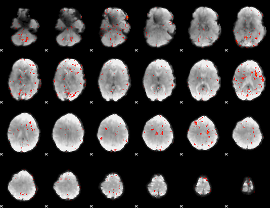 |
Correlation:0.13412
  |
Correlation:0.13412
  |
Correlation:0.13412
  |
Correlation:0.23053
  |
Correlation:0.13601
  |
Correlation:0.23053
  |
Correlation:0.12201
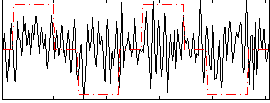 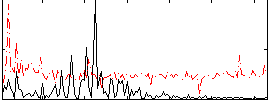 |
Cope: 04:"neg_PUNISH" BACK
|
GLM result group-wise
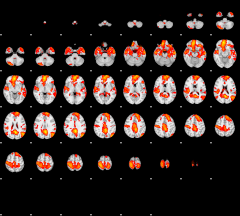 |
GLM binary result thresholded by 1
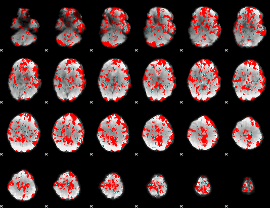 |
GLM binary result thresholded by 1.5
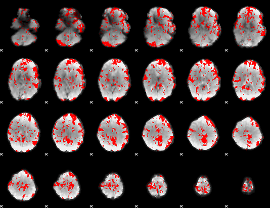 |
GLM binary result thresholded by 2
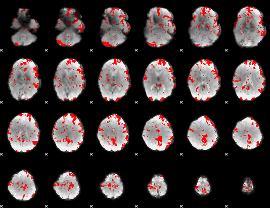 |
GLM binary result thresholded by 2.5
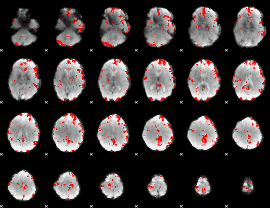 |
GLM binary result thresholded by 3
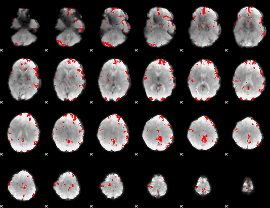 |
GLM binary result thresholded by 3.5
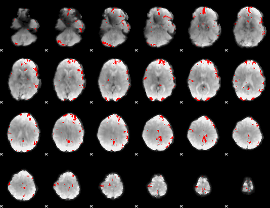 |
GLM binary result thresholded by 4
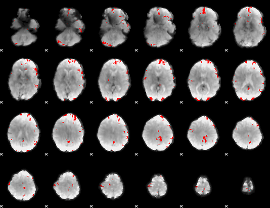 |
GLM Z-score result thresholded by 1
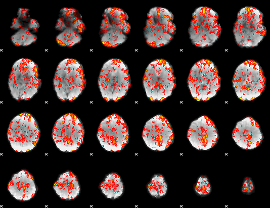 |
GLM Z-score result thresholded by 1.5
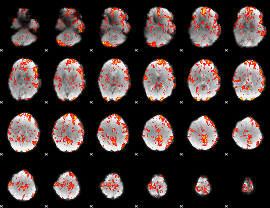 |
GLM Z-score result thresholded by 2
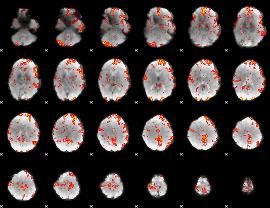 |
GLM Z-score result thresholded by 2.5
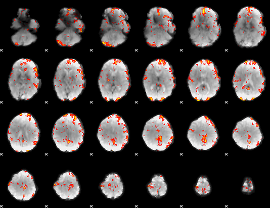 |
GLM Z-score result thresholded by 3
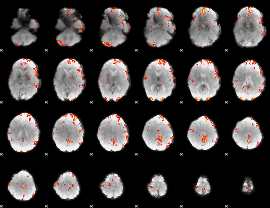 |
GLM Z-score result thresholded by 3.5
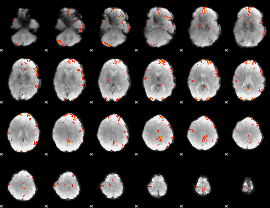 |
GLM Z-score result thresholded by 4
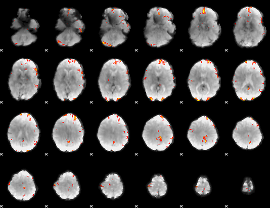 |
Component:351; Jaccard:0.15409
 |
Component:351; Jaccard:0.19683
 |
Component:351; Jaccard:0.24125
 |
Component:351; Jaccard:0.27287
 |
Component:351; Jaccard:0.27573
 |
Component:351; Jaccard:0.25436
 |
Component:351; Jaccard:0.21185
 |
Correlation:0.30943
  |
Correlation:0.30943
  |
Correlation:0.30943
  |
Correlation:0.30943
  |
Correlation:0.30943
  |
Correlation:0.30943
  |
Correlation:0.30943
  |
Component:180; Jaccard:0.13389
 |
Component:180; Jaccard:0.15986
 |
Component:180; Jaccard:0.18119
 |
Component:180; Jaccard:0.18992
 |
Component:180; Jaccard:0.17889
 |
Component:017; Jaccard:0.15771
 |
Component:017; Jaccard:0.12849
 |
Correlation:0.15843
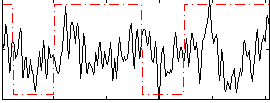 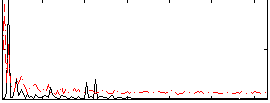 |
Correlation:0.15843
  |
Correlation:0.15843
  |
Correlation:0.15843
  |
Correlation:0.15843
  |
Correlation:0.37408
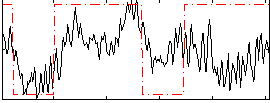 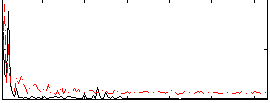 |
Correlation:0.37408
  |
Component:017; Jaccard:0.11164
 |
Component:017; Jaccard:0.13467
 |
Component:017; Jaccard:0.15544
 |
Component:017; Jaccard:0.16836
 |
Component:017; Jaccard:0.16877
 |
Component:180; Jaccard:0.15108
 |
Component:180; Jaccard:0.12225
 |
Correlation:0.37408
  |
Correlation:0.37408
  |
Correlation:0.37408
  |
Correlation:0.37408
  |
Correlation:0.37408
  |
Correlation:0.15843
  |
Correlation:0.15843
  |
Component:023; Jaccard:0.10814
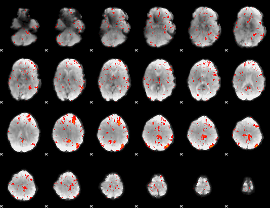 |
Component:023; Jaccard:0.121
 |
Component:023; Jaccard:0.12926
 |
Component:023; Jaccard:0.12736
 |
Component:056; Jaccard:0.12746
 |
Component:056; Jaccard:0.12311
 |
Component:056; Jaccard:0.1115
 |
Correlation:0.12046
  |
Correlation:0.12046
  |
Correlation:0.12046
  |
Correlation:0.12046
  |
Correlation:0.35142
 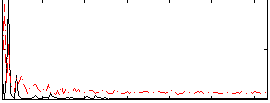 |
Correlation:0.35142
  |
Correlation:0.35142
  |
Component:328; Jaccard:0.1031
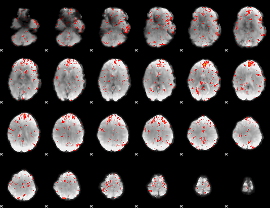 |
Component:328; Jaccard:0.10885
 |
Component:120; Jaccard:0.1188
 |
Component:056; Jaccard:0.12672
 |
Component:120; Jaccard:0.12003
 |
Component:120; Jaccard:0.10921
 |
Component:120; Jaccard:0.088247
 |
Correlation:-0.077093
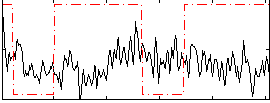 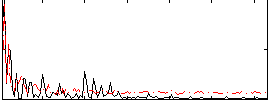 |
Correlation:-0.077093
  |
Correlation:0.062368
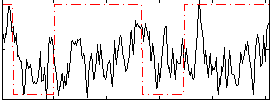 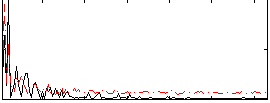 |
Correlation:0.35142
  |
Correlation:0.062368
  |
Correlation:0.062368
  |
Correlation:0.062368
  |
Component:357; Jaccard:0.097554
 |
Component:120; Jaccard:0.10803
 |
Component:056; Jaccard:0.11751
 |
Component:120; Jaccard:0.12305
 |
Component:023; Jaccard:0.1172
 |
Component:366; Jaccard:0.10064
 |
Component:366; Jaccard:0.085958
 |
Correlation:0.1221
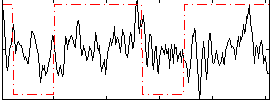 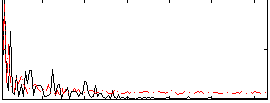 |
Correlation:0.062368
  |
Correlation:0.35142
  |
Correlation:0.062368
  |
Correlation:0.12046
  |
Correlation:0.18653
  |
Correlation:0.18653
  |
Component:366; Jaccard:0.097017
 |
Component:357; Jaccard:0.10802
 |
Component:366; Jaccard:0.11447
 |
Component:366; Jaccard:0.11727
 |
Component:366; Jaccard:0.11168
 |
Component:023; Jaccard:0.096349
 |
Component:291; Jaccard:0.073688
 |
Correlation:0.18653
  |
Correlation:0.1221
  |
Correlation:0.18653
  |
Correlation:0.18653
  |
Correlation:0.18653
  |
Correlation:0.12046
  |
Correlation:0.11637
 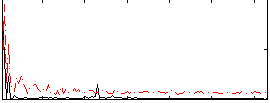 |
Component:120; Jaccard:0.094973
 |
Component:366; Jaccard:0.10743
 |
Component:357; Jaccard:0.112
 |
Component:291; Jaccard:0.11002
 |
Component:291; Jaccard:0.10668
 |
Component:291; Jaccard:0.093708
 |
Component:023; Jaccard:0.070586
 |
Correlation:0.062368
  |
Correlation:0.18653
  |
Correlation:0.1221
  |
Correlation:0.11637
  |
Correlation:0.11637
  |
Correlation:0.11637
  |
Correlation:0.12046
  |
Component:025; Jaccard:0.093589
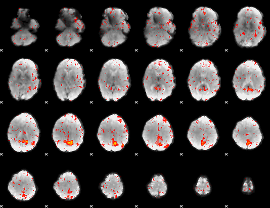 |
Component:056; Jaccard:0.10547
 |
Component:328; Jaccard:0.10836
 |
Component:357; Jaccard:0.1073
 |
Component:176; Jaccard:0.098998
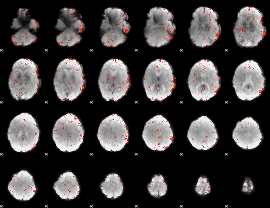 |
Component:176; Jaccard:0.088796
 |
Component:176; Jaccard:0.068951
 |
Correlation:0.18024
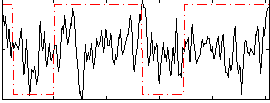  |
Correlation:0.35142
  |
Correlation:-0.077093
  |
Correlation:0.1221
  |
Correlation:0.2475
  |
Correlation:0.2475
  |
Correlation:0.2475
  |
Component:056; Jaccard:0.092183
 |
Component:025; Jaccard:0.099264
 |
Component:291; Jaccard:0.106
 |
Component:176; Jaccard:0.1043
 |
Component:105; Jaccard:0.095891
 |
Component:105; Jaccard:0.083853
 |
Component:310; Jaccard:0.064713
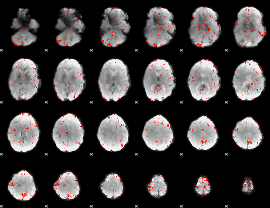 |
Correlation:0.35142
  |
Correlation:0.18024
  |
Correlation:0.11637
  |
Correlation:0.2475
  |
Correlation:0.13695
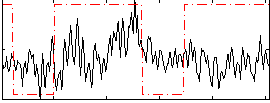 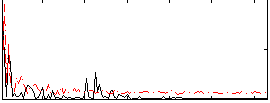 |
Correlation:0.13695
  |
Correlation:0.10906
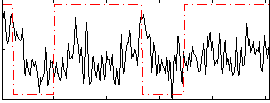 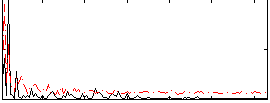 |
Cope: 05:"neg_REWARD" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
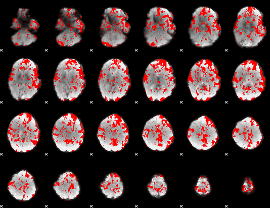 |
GLM binary result thresholded by 1.5
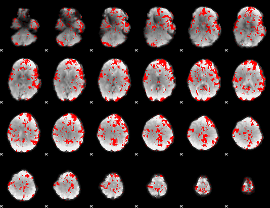 |
GLM binary result thresholded by 2
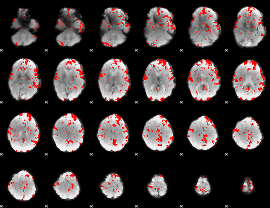 |
GLM binary result thresholded by 2.5
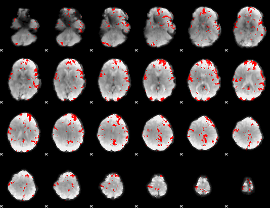 |
GLM binary result thresholded by 3
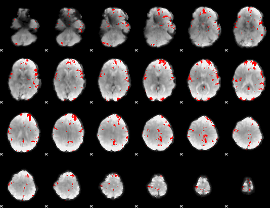 |
GLM binary result thresholded by 3.5
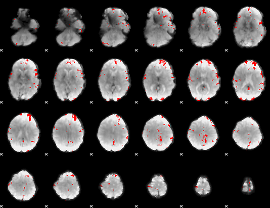 |
GLM binary result thresholded by 4
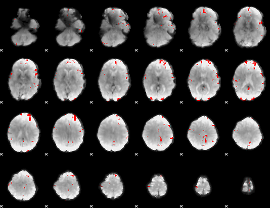 |
GLM Z-score result thresholded by 1
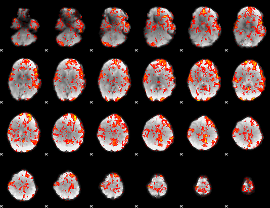 |
GLM Z-score result thresholded by 1.5
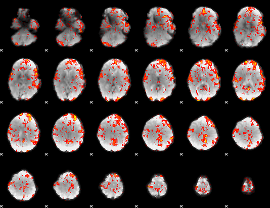 |
GLM Z-score result thresholded by 2
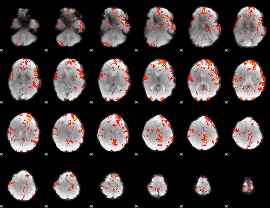 |
GLM Z-score result thresholded by 2.5
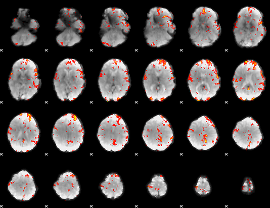 |
GLM Z-score result thresholded by 3
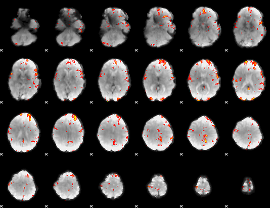 |
GLM Z-score result thresholded by 3.5
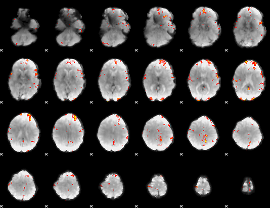 |
GLM Z-score result thresholded by 4
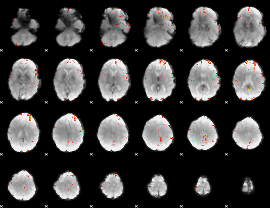 |
Component:180; Jaccard:0.13645
 |
Component:351; Jaccard:0.16556
 |
Component:351; Jaccard:0.19221
 |
Component:351; Jaccard:0.20111
 |
Component:351; Jaccard:0.18976
 |
Component:351; Jaccard:0.15909
 |
Component:351; Jaccard:0.12006
 |
Correlation:0.21061
  |
Correlation:0.15236
  |
Correlation:0.15236
  |
Correlation:0.15236
  |
Correlation:0.15236
  |
Correlation:0.15236
  |
Correlation:0.15236
  |
Component:351; Jaccard:0.13587
 |
Component:180; Jaccard:0.16138
 |
Component:180; Jaccard:0.17889
 |
Component:180; Jaccard:0.18383
 |
Component:180; Jaccard:0.16888
 |
Component:180; Jaccard:0.13954
 |
Component:056; Jaccard:0.1103
 |
Correlation:0.15236
  |
Correlation:0.21061
  |
Correlation:0.21061
  |
Correlation:0.21061
  |
Correlation:0.21061
  |
Correlation:0.21061
  |
Correlation:0.19177
  |
Component:348; Jaccard:0.10919
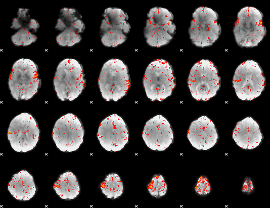 |
Component:056; Jaccard:0.12682
 |
Component:056; Jaccard:0.14611
 |
Component:056; Jaccard:0.15682
 |
Component:056; Jaccard:0.15148
 |
Component:056; Jaccard:0.13693
 |
Component:180; Jaccard:0.10313
 |
Correlation:0.16275
  |
Correlation:0.19177
  |
Correlation:0.19177
  |
Correlation:0.19177
  |
Correlation:0.19177
  |
Correlation:0.19177
  |
Correlation:0.21061
  |
Component:056; Jaccard:0.10667
 |
Component:348; Jaccard:0.11668
 |
Component:120; Jaccard:0.12399
 |
Component:120; Jaccard:0.12596
 |
Component:120; Jaccard:0.1163
 |
Component:120; Jaccard:0.10152
 |
Component:120; Jaccard:0.084301
 |
Correlation:0.19177
  |
Correlation:0.16275
  |
Correlation:0.251
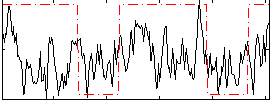  |
Correlation:0.251
  |
Correlation:0.251
  |
Correlation:0.251
  |
Correlation:0.251
  |
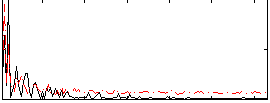
Component:375; Jaccard:0.097545
 |
Component:120; Jaccard:0.11294
 |
Component:348; Jaccard:0.11718
 |
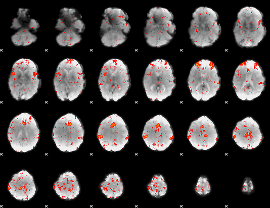
Component:059; Jaccard:0.11431
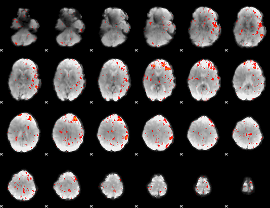 |
Component:059; Jaccard:0.10626
 |
Component:059; Jaccard:0.092736
 |
Component:059; Jaccard:0.071744
 |
Correlation:0.29139
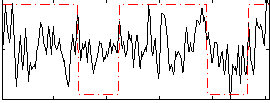 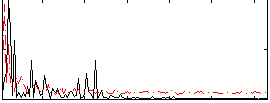 |
Correlation:0.251
  |
Correlation:0.16275
  |
Correlation:0.26445
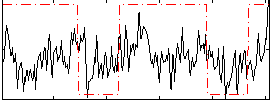 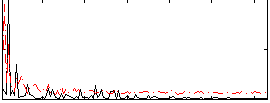 |
Correlation:0.26445
  |
Correlation:0.26445
  |
Correlation:0.26445
  |
Component:366; Jaccard:0.097505
 |
Component:366; Jaccard:0.10701
 |
Component:059; Jaccard:0.11342
 |
Component:348; Jaccard:0.11091
 |
Component:086; Jaccard:0.10516
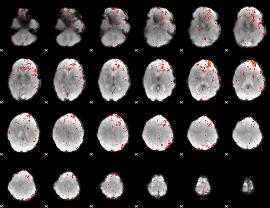 |
Component:086; Jaccard:0.090335
 |
Component:086; Jaccard:0.06946
 |
Correlation:0.10002
  |
Correlation:0.10002
  |
Correlation:0.26445
  |
Correlation:0.16275
  |
Correlation:0.47236
  |
Correlation:0.47236
  |
Correlation:0.47236
  |
Component:120; Jaccard:0.096973
 |
Component:059; Jaccard:0.10444
 |
Component:366; Jaccard:0.11256
 |
Component:086; Jaccard:0.11053
 |
Component:366; Jaccard:0.099972
 |
Component:366; Jaccard:0.083014
 |
Component:366; Jaccard:0.0638
 |
Correlation:0.251
  |
Correlation:0.26445
  |
Correlation:0.10002
  |
Correlation:0.47236
  |
Correlation:0.10002
  |
Correlation:0.10002
  |
Correlation:0.10002
  |
Component:059; Jaccard:0.090836
 |
Component:375; Jaccard:0.10311
 |
Component:086; Jaccard:0.10763
 |
Component:366; Jaccard:0.11041
 |
Component:348; Jaccard:0.095461
 |
Component:017; Jaccard:0.080929
 |
Component:017; Jaccard:0.063133
 |
Correlation:0.26445
  |
Correlation:0.29139
  |
Correlation:0.47236
  |
Correlation:0.10002
  |
Correlation:0.16275
  |
Correlation:0.078181
  |
Correlation:0.078181
  |
Component:167; Jaccard:0.086962
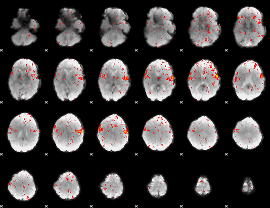 |
Component:167; Jaccard:0.098239
 |
Component:375; Jaccard:0.10639
 |
Component:375; Jaccard:0.10184
 |
Component:017; Jaccard:0.094445
 |
Component:348; Jaccard:0.075379
 |
Component:348; Jaccard:0.056232
 |
Correlation:0.38789
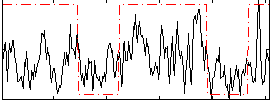 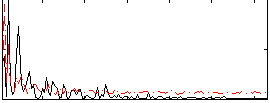 |
Correlation:0.38789
  |
Correlation:0.29139
  |
Correlation:0.29139
  |
Correlation:0.078181
  |
Correlation:0.16275
  |
Correlation:0.16275
  |
Component:352; Jaccard:0.086362
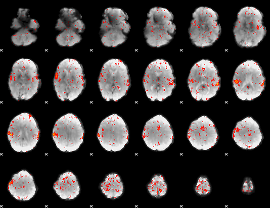 |
Component:086; Jaccard:0.097429
 |
Component:167; Jaccard:0.10244
 |
Component:017; Jaccard:0.099771
 |
Component:375; Jaccard:0.089446
 |
Component:375; Jaccard:0.070686
 |
Component:375; Jaccard:0.052485
 |
Correlation:0.15011
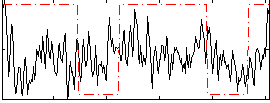 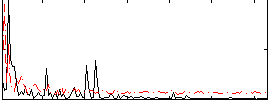 |
Correlation:0.47236
  |
Correlation:0.38789
  |
Correlation:0.078181
  |
Correlation:0.29139
  |
Correlation:0.29139
  |
Correlation:0.29139
  |
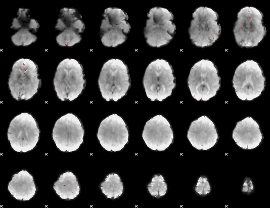
Cope: 06:"REWARD-PUNISH" BACK
|
GLM result group-wise
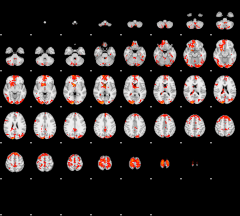 |
GLM binary result thresholded by 1
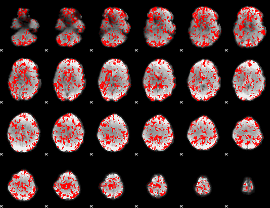 |
GLM binary result thresholded by 1.5
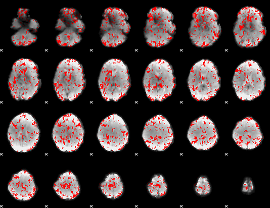 |
GLM binary result thresholded by 2
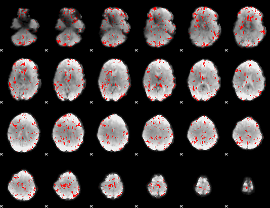 |
GLM binary result thresholded by 2.5
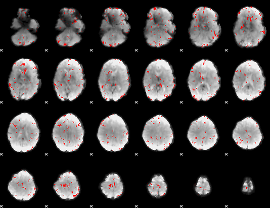 |
GLM binary result thresholded by 3
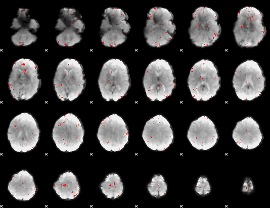 |
GLM binary result thresholded by 3.5
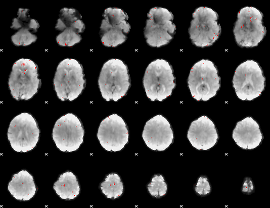 |
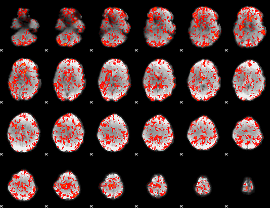
GLM binary result thresholded by 4
 |
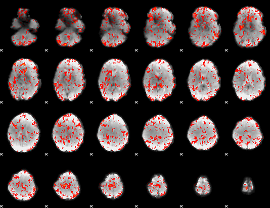
GLM Z-score result thresholded by 1
 |
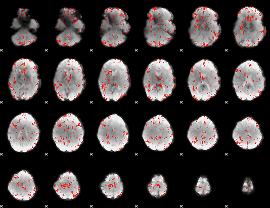
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
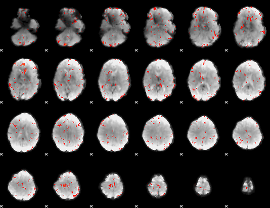 |
GLM Z-score result thresholded by 3
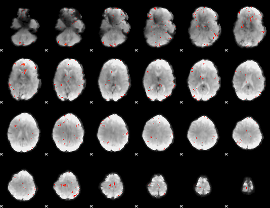 |
GLM Z-score result thresholded by 3.5
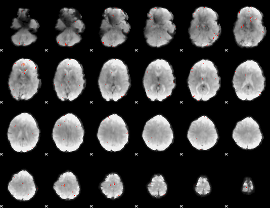 |
GLM Z-score result thresholded by 4
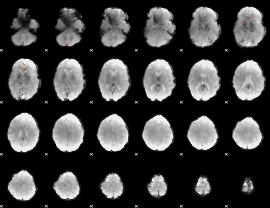 |
Component:357; Jaccard:0.10456
 |
Component:357; Jaccard:0.1166
 |
Component:357; Jaccard:0.11189
 |
Component:357; Jaccard:0.084036
 |
Component:357; Jaccard:0.049263
 |
Component:357; Jaccard:0.023445
 |
Component:356; Jaccard:0.008859
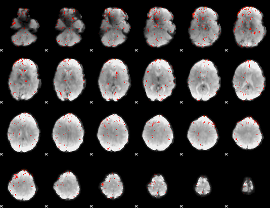 |
Correlation:0.10377
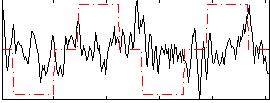 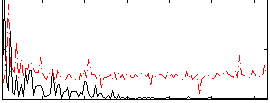 |
Correlation:0.10377
  |
Correlation:0.10377
  |
Correlation:0.10377
  |
Correlation:0.10377
  |
Correlation:0.10377
  |
Correlation:0.55408
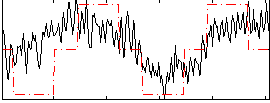 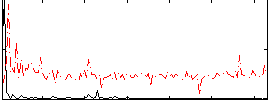 |
Component:325; Jaccard:0.085475
 |
Component:017; Jaccard:0.086721
 |
Component:017; Jaccard:0.083568
 |
Component:356; Jaccard:0.067984
 |
Component:356; Jaccard:0.043528
 |
Component:356; Jaccard:0.020631
 |
Component:296; Jaccard:0.00822
 |
Correlation:0.072307
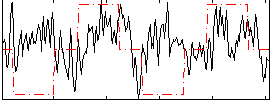 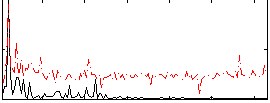 |
Correlation:0.17402
  |
Correlation:0.17402
  |
Correlation:0.55408
  |
Correlation:0.55408
  |
Correlation:0.55408
  |
Correlation:0.23414
  |
Component:255; Jaccard:0.082599
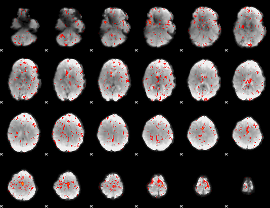 |
Component:325; Jaccard:0.085996
 |
Component:325; Jaccard:0.07857
 |
Component:017; Jaccard:0.067037
 |
Component:017; Jaccard:0.03852
 |
Component:017; Jaccard:0.016768
 |
Component:357; Jaccard:0.008078
 |
Correlation:0.020927
  |
Correlation:0.072307
  |
Correlation:0.072307
  |
Correlation:0.17402
  |
Correlation:0.17402
  |
Correlation:0.17402
  |
Correlation:0.10377
  |
Component:093; Jaccard:0.080472
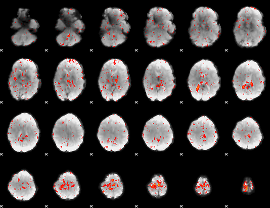 |
Component:255; Jaccard:0.083848
 |
Component:356; Jaccard:0.07774
 |
Component:176; Jaccard:0.05833
 |
Component:027; Jaccard:0.031837
 |
Component:027; Jaccard:0.016385
 |
Component:017; Jaccard:0.006693
 |
Correlation:0.051012
  |
Correlation:0.020927
  |
Correlation:0.55408
  |
Correlation:0.15281
  |
Correlation:0.080401
  |
Correlation:0.080401
  |
Correlation:0.17402
  |
Component:284; Jaccard:0.080095
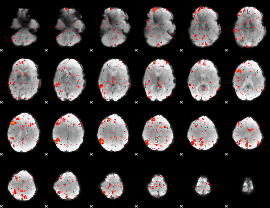 |
Component:093; Jaccard:0.0838
 |
Component:071; Jaccard:0.075443
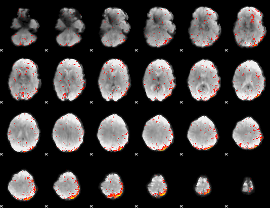 |
Component:325; Jaccard:0.055604
 |
Component:071; Jaccard:0.031498
 |
Component:296; Jaccard:0.015194
 |
Component:027; Jaccard:0.00665
 |
Correlation:0.084913
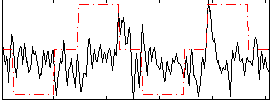 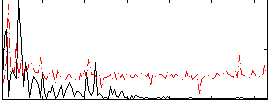 |
Correlation:0.051012
  |
Correlation:-0.055544
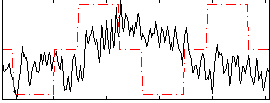 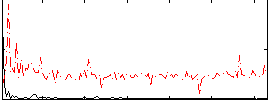 |
Correlation:0.072307
  |
Correlation:-0.055544
  |
Correlation:0.23414
  |
Correlation:0.080401
  |
Component:233; Jaccard:0.079064
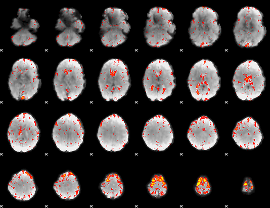 |
Component:284; Jaccard:0.081565
 |
Component:255; Jaccard:0.074682
 |
Component:071; Jaccard:0.052917
 |
Component:093; Jaccard:0.03043
 |
Component:176; Jaccard:0.013664
 |
Component:255; Jaccard:0.005477
 |
Correlation:0.10062
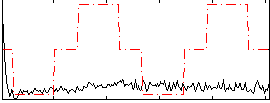 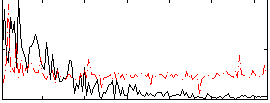 |
Correlation:0.084913
  |
Correlation:0.020927
  |
Correlation:-0.055544
  |
Correlation:0.051012
  |
Correlation:0.15281
  |
Correlation:0.020927
  |
Component:027; Jaccard:0.078445
 |
Component:071; Jaccard:0.080815
 |
Component:093; Jaccard:0.074151
 |
Component:093; Jaccard:0.052767
 |
Component:176; Jaccard:0.029962
 |
Component:143; Jaccard:0.013331
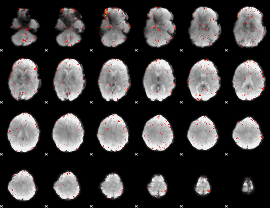 |
Component:269; Jaccard:0.004751
 |
Correlation:0.080401
  |
Correlation:-0.055544
  |
Correlation:0.051012
  |
Correlation:0.051012
  |
Correlation:0.15281
  |
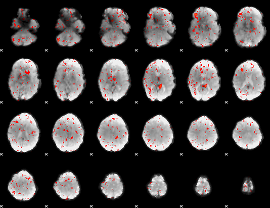
Correlation:0.38125
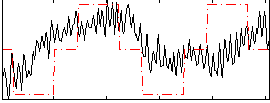 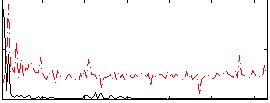 |
Correlation:0.099022
  |
Component:157; Jaccard:0.078084
 |
Component:027; Jaccard:0.07888
 |
Component:176; Jaccard:0.073725
 |
Component:027; Jaccard:0.052308
 |
Component:255; Jaccard:0.029072
 |
Component:093; Jaccard:0.012583
 |
Component:108; Jaccard:0.004333
 |
Correlation:0.11705
  |
Correlation:0.080401
  |
Correlation:0.15281
  |
Correlation:0.080401
  |
Correlation:0.020927
  |
Correlation:0.051012
  |
Correlation:0.090176
  |
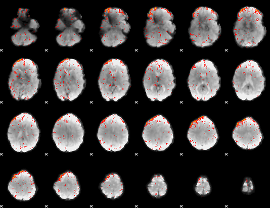
Component:017; Jaccard:0.077143
 |
Component:157; Jaccard:0.078113
 |
Component:027; Jaccard:0.070626
 |
Component:255; Jaccard:0.051889
 |
Component:325; Jaccard:0.028995
 |
Component:255; Jaccard:0.012029
 |
Component:168; Jaccard:0.004169
 |
Correlation:0.17402
  |
Correlation:0.11705
  |
Correlation:0.080401
  |
Correlation:0.020927
  |
Correlation:0.072307
  |
Correlation:0.020927
  |
Correlation:0.042452
  |
Component:071; Jaccard:0.07673
 |
Component:176; Jaccard:0.076777
 |
Component:284; Jaccard:0.069591
 |
Component:284; Jaccard:0.047982
 |
Component:143; Jaccard:0.028236
 |
Component:071; Jaccard:0.011998
 |
Component:143; Jaccard:0.004147
 |
Correlation:-0.055544
  |
Correlation:0.15281
  |
Correlation:0.084913
  |
Correlation:0.084913
  |
Correlation:0.38125
  |
Correlation:-0.055544
  |
Correlation:0.38125
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































