Task name: LANGUAGE, TR=0.72, totally 316 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"MATH" BACK
|
GLM result group-wise
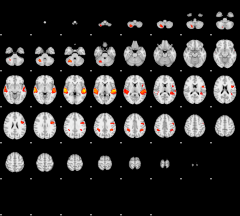 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
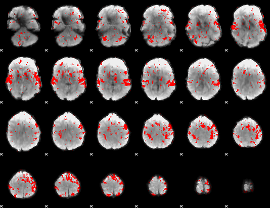 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
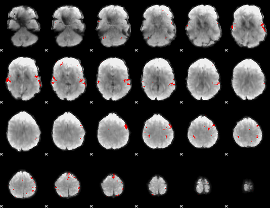 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
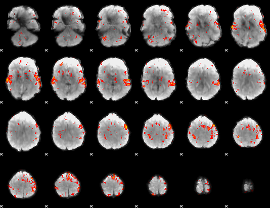 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:125; Jaccard:0.15053
 |
Component:125; Jaccard:0.16799
 |
Component:392; Jaccard:0.17662
 |
Component:392; Jaccard:0.16083
 |
Component:392; Jaccard:0.1297
 |
Component:392; Jaccard:0.093911
 |
Component:392; Jaccard:0.066836
 |
Correlation:0.29059
  |
Correlation:0.29059
  |
Correlation:0.079822
  |
Correlation:0.079822
  |
Correlation:0.079822
  |
Correlation:0.079822
  |
Correlation:0.079822
  |
Component:374; Jaccard:0.14073
 |
Component:392; Jaccard:0.16556
 |
Component:374; Jaccard:0.16198
 |
Component:374; Jaccard:0.14357
 |
Component:374; Jaccard:0.11383
 |
Component:374; Jaccard:0.082162
 |
Component:374; Jaccard:0.05922
 |
Correlation:-0.30459
  |
Correlation:0.079822
  |
Correlation:-0.30459
  |
Correlation:-0.30459
  |
Correlation:-0.30459
  |
Correlation:-0.30459
  |
Correlation:-0.30459
  |
Component:392; Jaccard:0.13858
 |
Component:374; Jaccard:0.16003
 |
Component:125; Jaccard:0.16089
 |
Component:125; Jaccard:0.13434
 |
Component:125; Jaccard:0.10055
 |
Component:125; Jaccard:0.067599
 |
Component:341; Jaccard:0.0462
 |
Correlation:0.079822
  |
Correlation:-0.30459
  |
Correlation:0.29059
  |
Correlation:0.29059
  |
Correlation:0.29059
  |
Correlation:0.29059
  |
Correlation:-0.10334
  |
Component:045; Jaccard:0.11424
 |
Component:341; Jaccard:0.13009
 |
Component:341; Jaccard:0.13336
 |
Component:341; Jaccard:0.11806
 |
Component:341; Jaccard:0.095406
 |
Component:341; Jaccard:0.067001
 |
Component:125; Jaccard:0.044503
 |
Correlation:0.50407
  |
Correlation:-0.10334
  |
Correlation:-0.10334
  |
Correlation:-0.10334
  |
Correlation:-0.10334
  |
Correlation:-0.10334
  |
Correlation:0.29059
  |
Component:341; Jaccard:0.11369
 |
Component:045; Jaccard:0.12119
 |
Component:045; Jaccard:0.11526
 |
Component:045; Jaccard:0.09428
 |
Component:045; Jaccard:0.067258
 |
Component:103; Jaccard:0.040361
 |
Component:103; Jaccard:0.025589
 |
Correlation:-0.10334
  |
Correlation:0.50407
  |
Correlation:0.50407
  |
Correlation:0.50407
  |
Correlation:0.50407
  |
Correlation:0.23393
  |
Correlation:0.23393
  |
Component:173; Jaccard:0.1111
 |
Component:103; Jaccard:0.11192
 |
Component:103; Jaccard:0.10538
 |
Component:103; Jaccard:0.087092
 |
Component:103; Jaccard:0.065752
 |
Component:045; Jaccard:0.0386
 |
Component:063; Jaccard:0.025529
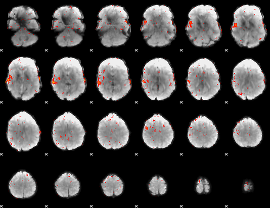 |
Correlation:0.16485
  |
Correlation:0.23393
  |
Correlation:0.23393
  |
Correlation:0.23393
  |
Correlation:0.23393
  |
Correlation:0.50407
  |
Correlation:-0.32287
  |
Component:150; Jaccard:0.10581
 |
Component:150; Jaccard:0.10619
 |
Component:150; Jaccard:0.08868
 |
Component:150; Jaccard:0.06328
 |
Component:063; Jaccard:0.048084
 |
Component:063; Jaccard:0.036909
 |
Component:045; Jaccard:0.021028
 |
Correlation:0.68368
  |
Correlation:0.68368
  |
Correlation:0.68368
  |
Correlation:0.68368
  |
Correlation:-0.32287
  |
Correlation:-0.32287
  |
Correlation:0.50407
  |
Component:103; Jaccard:0.10311
 |
Component:173; Jaccard:0.10487
 |
Component:173; Jaccard:0.084265
 |
Component:173; Jaccard:0.059939
 |
Component:364; Jaccard:0.042771
 |
Component:364; Jaccard:0.030152
 |
Component:269; Jaccard:0.020557
 |
Correlation:0.23393
  |
Correlation:0.16485
  |
Correlation:0.16485
  |
Correlation:0.16485
  |
Correlation:-0.15136
  |
Correlation:-0.15136
  |
Correlation:0.10217
  |
Component:272; Jaccard:0.085591
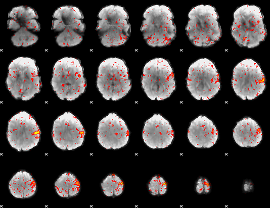 |
Component:168; Jaccard:0.077976
 |
Component:168; Jaccard:0.071253
 |
Component:364; Jaccard:0.05736
 |
Component:150; Jaccard:0.042529
 |
Component:269; Jaccard:0.029082
 |
Component:364; Jaccard:0.019681
 |
Correlation:0.18592
  |
Correlation:0.11584
  |
Correlation:0.11584
  |
Correlation:-0.15136
  |
Correlation:0.68368
  |
Correlation:0.10217
  |
Correlation:-0.15136
  |
Component:269; Jaccard:0.079514
 |
Component:272; Jaccard:0.077217
 |
Component:364; Jaccard:0.068003
 |
Component:063; Jaccard:0.057131
 |
Component:269; Jaccard:0.040483
 |
Component:150; Jaccard:0.026764
 |
Component:150; Jaccard:0.016618
 |
Correlation:0.10217
  |
Correlation:0.18592
  |
Correlation:-0.15136
  |
Correlation:-0.32287
  |
Correlation:0.10217
  |
Correlation:0.68368
  |
Correlation:0.68368
  |
Cope: 02:"STORY" BACK
|
GLM result group-wise
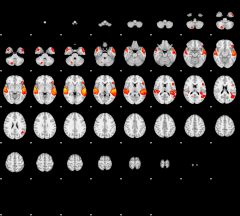 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
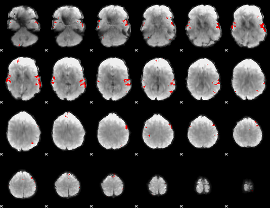 |
GLM Z-score result thresholded by 1
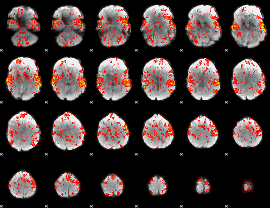 |
GLM Z-score result thresholded by 1.5
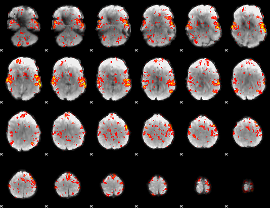 |
GLM Z-score result thresholded by 2
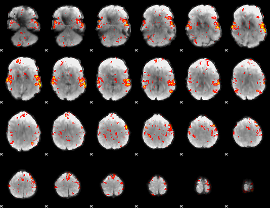 |
GLM Z-score result thresholded by 2.5
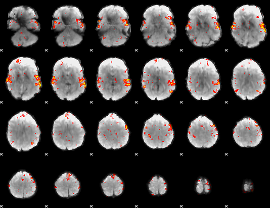 |
GLM Z-score result thresholded by 3
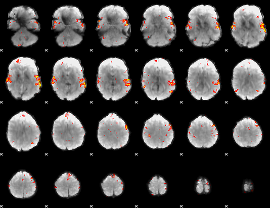 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
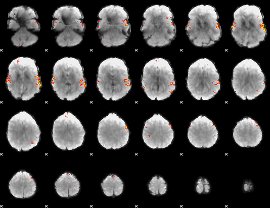 |
Component:374; Jaccard:0.18372
 |
Component:374; Jaccard:0.23033
 |
Component:374; Jaccard:0.26234
 |
Component:374; Jaccard:0.27184
 |
Component:374; Jaccard:0.2487
 |
Component:374; Jaccard:0.21063
 |
Component:374; Jaccard:0.16616
 |
Correlation:0.2592
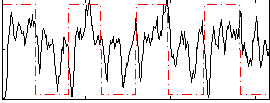 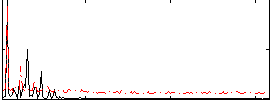 |
Correlation:0.2592
  |
Correlation:0.2592
  |
Correlation:0.2592
  |
Correlation:0.2592
  |
Correlation:0.2592
  |
Correlation:0.2592
  |
Component:129; Jaccard:0.12229
 |
Component:129; Jaccard:0.1395
 |
Component:341; Jaccard:0.14604
 |
Component:341; Jaccard:0.14759
 |
Component:341; Jaccard:0.13526
 |
Component:341; Jaccard:0.11395
 |
Component:341; Jaccard:0.088262
 |
Correlation:0.46506
  |
Correlation:0.46506
  |
Correlation:0.075551
  |
Correlation:0.075551
  |
Correlation:0.075551
  |
Correlation:0.075551
  |
Correlation:0.075551
  |
Component:222; Jaccard:0.12212
 |
Component:222; Jaccard:0.135
 |
Component:129; Jaccard:0.14302
 |
Component:129; Jaccard:0.13548
 |
Component:392; Jaccard:0.12528
 |
Component:392; Jaccard:0.10801
 |
Component:392; Jaccard:0.087908
 |
Correlation:0.37595
  |
Correlation:0.37595
  |
Correlation:0.46506
  |
Correlation:0.46506
  |
Correlation:-0.099693
  |
Correlation:-0.099693
  |
Correlation:-0.099693
  |
Component:341; Jaccard:0.11659
 |
Component:341; Jaccard:0.13492
 |
Component:392; Jaccard:0.13416
 |
Component:392; Jaccard:0.134
 |
Component:129; Jaccard:0.11346
 |
Component:129; Jaccard:0.087097
 |
Component:129; Jaccard:0.064254
 |
Correlation:0.075551
  |
Correlation:0.075551
  |
Correlation:-0.099693
  |
Correlation:-0.099693
  |
Correlation:0.46506
  |
Correlation:0.46506
  |
Correlation:0.46506
  |
Component:392; Jaccard:0.10558
 |
Component:392; Jaccard:0.1245
 |
Component:222; Jaccard:0.13207
 |
Component:222; Jaccard:0.11474
 |
Component:026; Jaccard:0.092634
 |
Component:026; Jaccard:0.072705
 |
Component:026; Jaccard:0.055099
 |
Correlation:-0.099693
  |
Correlation:-0.099693
  |
Correlation:0.37595
  |
Correlation:0.37595
  |
Correlation:0.63281
  |
Correlation:0.63281
  |
Correlation:0.63281
  |
Component:241; Jaccard:0.098503
 |
Component:026; Jaccard:0.10101
 |
Component:026; Jaccard:0.10883
 |
Component:026; Jaccard:0.10742
 |
Component:222; Jaccard:0.086438
 |
Component:063; Jaccard:0.066595
 |
Component:063; Jaccard:0.051631
 |
Correlation:0.3082
  |
Correlation:0.63281
  |
Correlation:0.63281
  |
Correlation:0.63281
  |
Correlation:0.37595
  |
Correlation:0.2917
  |
Correlation:0.2917
  |
Component:173; Jaccard:0.097767
 |
Component:241; Jaccard:0.10018
 |
Component:241; Jaccard:0.094498
 |
Component:181; Jaccard:0.086296
 |
Component:063; Jaccard:0.073707
 |
Component:222; Jaccard:0.056196
 |
Component:310; Jaccard:0.045346
 |
Correlation:-0.20186
  |
Correlation:0.3082
  |
Correlation:0.3082
  |
Correlation:0.2796
  |
Correlation:0.2917
  |
Correlation:0.37595
  |
Correlation:0.25695
  |
Component:125; Jaccard:0.096395
 |
Component:125; Jaccard:0.099581
 |
Component:125; Jaccard:0.094234
 |
Component:241; Jaccard:0.081804
 |
Component:181; Jaccard:0.072739
 |
Component:181; Jaccard:0.054467
 |
Component:181; Jaccard:0.041928
 |
Correlation:-0.31084
  |
Correlation:-0.31084
  |
Correlation:-0.31084
  |
Correlation:0.3082
  |
Correlation:0.2796
  |
Correlation:0.2796
  |
Correlation:0.2796
  |
Component:026; Jaccard:0.087828
 |
Component:181; Jaccard:0.092048
 |
Component:181; Jaccard:0.093068
 |
Component:125; Jaccard:0.080383
 |
Component:125; Jaccard:0.065502
 |
Component:310; Jaccard:0.051294
 |
Component:269; Jaccard:0.038299
 |
Correlation:0.63281
  |
Correlation:0.2796
  |
Correlation:0.2796
  |
Correlation:-0.31084
  |
Correlation:-0.31084
  |
Correlation:0.25695
  |
Correlation:-0.16446
  |
Component:269; Jaccard:0.087248
 |
Component:173; Jaccard:0.091552
 |
Component:364; Jaccard:0.086783
 |
Component:063; Jaccard:0.075941
 |
Component:241; Jaccard:0.065442
 |
Component:269; Jaccard:0.049355
 |
Component:222; Jaccard:0.037294
 |
Correlation:-0.16446
  |
Correlation:-0.20186
  |
Correlation:0.20984
  |
Correlation:0.2917
  |
Correlation:0.3082
  |
Correlation:-0.16446
  |
Correlation:0.37595
  |
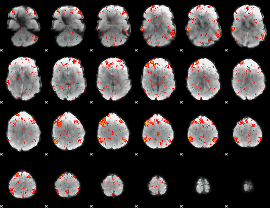
Cope: 03:"MATH-STORY" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
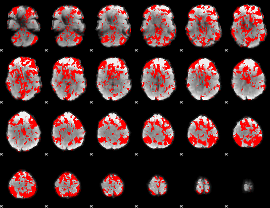 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
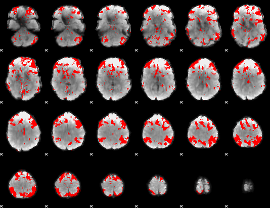 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
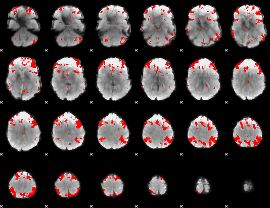 |
GLM binary result thresholded by 4
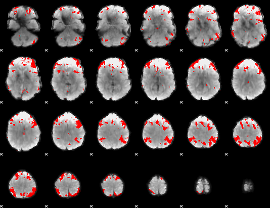 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
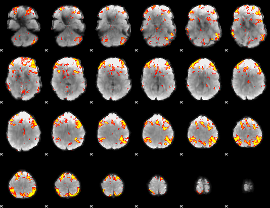 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
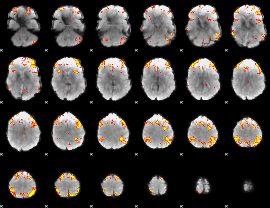 |
Component:306; Jaccard:0.16349
 |
Component:306; Jaccard:0.19609
 |
Component:306; Jaccard:0.23133
 |
Component:306; Jaccard:0.26088
 |
Component:306; Jaccard:0.28287
 |
Component:306; Jaccard:0.29403
 |
Component:306; Jaccard:0.29651
 |
Correlation:0.58021
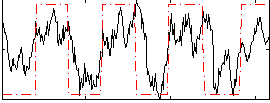 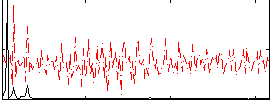 |
Correlation:0.58021
  |
Correlation:0.58021
  |
Correlation:0.58021
  |
Correlation:0.58021
  |
Correlation:0.58021
  |
Correlation:0.58021
  |
Component:263; Jaccard:0.15756
 |
Component:370; Jaccard:0.18445
 |
Component:370; Jaccard:0.21198
 |
Component:370; Jaccard:0.2374
 |
Component:370; Jaccard:0.25602
 |
Component:370; Jaccard:0.26768
 |
Component:370; Jaccard:0.27049
 |
Correlation:0.47081
  |
Correlation:0.51621
  |
Correlation:0.51621
  |
Correlation:0.51621
  |
Correlation:0.51621
  |
Correlation:0.51621
  |
Correlation:0.51621
  |
Component:370; Jaccard:0.15693
 |
Component:263; Jaccard:0.18254
 |
Component:263; Jaccard:0.20745
 |
Component:263; Jaccard:0.22949
 |
Component:263; Jaccard:0.24419
 |
Component:045; Jaccard:0.25667
 |
Component:045; Jaccard:0.26955
 |
Correlation:0.51621
  |
Correlation:0.47081
  |
Correlation:0.47081
  |
Correlation:0.47081
  |
Correlation:0.47081
  |
Correlation:0.53205
  |
Correlation:0.53205
  |
Component:115; Jaccard:0.15319
 |
Component:115; Jaccard:0.17251
 |
Component:115; Jaccard:0.19058
 |
Component:045; Jaccard:0.21508
 |
Component:045; Jaccard:0.23875
 |
Component:263; Jaccard:0.25314
 |
Component:263; Jaccard:0.24946
 |
Correlation:0.57052
  |
Correlation:0.57052
  |
Correlation:0.57052
  |
Correlation:0.53205
  |
Correlation:0.53205
  |
Correlation:0.47081
  |
Correlation:0.47081
  |
Component:326; Jaccard:0.14248
 |
Component:045; Jaccard:0.15831
 |
Component:045; Jaccard:0.18631
 |
Component:115; Jaccard:0.20279
 |
Component:115; Jaccard:0.20777
 |
Component:115; Jaccard:0.20445
 |
Component:354; Jaccard:0.20466
 |
Correlation:0.22123
  |
Correlation:0.53205
  |
Correlation:0.53205
  |
Correlation:0.57052
  |
Correlation:0.57052
  |
Correlation:0.57052
  |
Correlation:0.5488
  |
Component:218; Jaccard:0.13738
 |
Component:326; Jaccard:0.15523
 |
Component:150; Jaccard:0.174
 |
Component:150; Jaccard:0.18855
 |
Component:150; Jaccard:0.19634
 |
Component:354; Jaccard:0.20192
 |
Component:115; Jaccard:0.19489
 |
Correlation:0.5424
  |
Correlation:0.22123
  |
Correlation:0.71728
  |
Correlation:0.71728
  |
Correlation:0.71728
  |
Correlation:0.5488
  |
Correlation:0.57052
  |
Component:109; Jaccard:0.13662
 |
Component:150; Jaccard:0.15518
 |
Component:218; Jaccard:0.16819
 |
Component:354; Jaccard:0.18368
 |
Component:354; Jaccard:0.19578
 |
Component:150; Jaccard:0.19436
 |
Component:150; Jaccard:0.18782
 |
Correlation:0.31313
  |
Correlation:0.71728
  |
Correlation:0.5424
  |
Correlation:0.5488
  |
Correlation:0.5488
  |
Correlation:0.71728
  |
Correlation:0.71728
  |
Component:150; Jaccard:0.13436
 |
Component:218; Jaccard:0.15384
 |
Component:326; Jaccard:0.16728
 |
Component:218; Jaccard:0.17701
 |
Component:218; Jaccard:0.18146
 |
Component:215; Jaccard:0.18333
 |
Component:215; Jaccard:0.17992
 |
Correlation:0.71728
  |
Correlation:0.5424
  |
Correlation:0.22123
  |
Correlation:0.5424
  |
Correlation:0.5424
  |
Correlation:0.55234
  |
Correlation:0.55234
  |
Component:045; Jaccard:0.13169
 |
Component:109; Jaccard:0.14955
 |
Component:354; Jaccard:0.16526
 |
Component:326; Jaccard:0.17403
 |
Component:215; Jaccard:0.18041
 |
Component:218; Jaccard:0.17819
 |
Component:218; Jaccard:0.16976
 |
Correlation:0.53205
  |
Correlation:0.31313
  |
Correlation:0.5488
  |
Correlation:0.22123
  |
Correlation:0.55234
  |
Correlation:0.5424
  |
Correlation:0.5424
  |
Component:215; Jaccard:0.12618
 |
Component:354; Jaccard:0.14371
 |
Component:109; Jaccard:0.16115
 |
Component:215; Jaccard:0.17204
 |
Component:109; Jaccard:0.17378
 |
Component:109; Jaccard:0.17157
 |
Component:109; Jaccard:0.16486
 |
Correlation:0.55234
  |
Correlation:0.5488
  |
Correlation:0.31313
  |
Correlation:0.55234
  |
Correlation:0.31313
  |
Correlation:0.31313
  |
Correlation:0.31313
  |
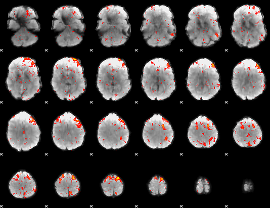
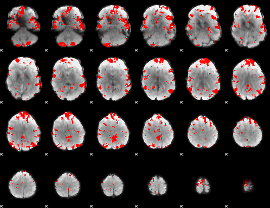
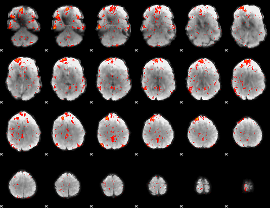
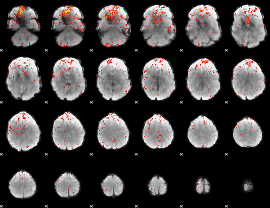
Cope: 04:"STORY-MATH" BACK
|
GLM result group-wise
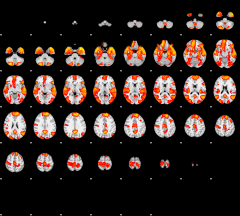 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
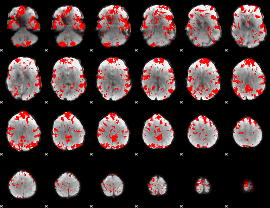 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
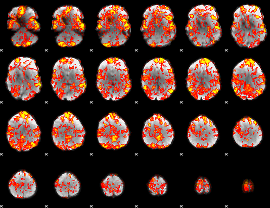 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
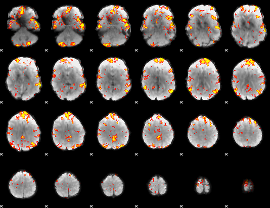 |
GLM Z-score result thresholded by 4
 |
Component:241; Jaccard:0.12491
 |
Component:241; Jaccard:0.14682
 |
Component:241; Jaccard:0.17095
 |
Component:241; Jaccard:0.19661
 |
Component:129; Jaccard:0.23427
 |
Component:129; Jaccard:0.27334
 |
Component:129; Jaccard:0.30781
 |
Correlation:0.33483
  |
Correlation:0.33483
  |
Correlation:0.33483
  |
Correlation:0.33483
  |
Correlation:0.48332
  |
Correlation:0.48332
  |
Correlation:0.48332
  |
Component:250; Jaccard:0.10848
 |
Component:129; Jaccard:0.12743
 |
Component:129; Jaccard:0.15838
 |
Component:129; Jaccard:0.19509
 |
Component:241; Jaccard:0.2174
 |
Component:241; Jaccard:0.23253
 |
Component:241; Jaccard:0.2368
 |
Correlation:0.33912
  |
Correlation:0.48332
  |
Correlation:0.48332
  |
Correlation:0.48332
  |
Correlation:0.33483
  |
Correlation:0.33483
  |
Correlation:0.33483
  |
Component:158; Jaccard:0.10635
 |
Component:158; Jaccard:0.12582
 |
Component:203; Jaccard:0.14758
 |
Component:158; Jaccard:0.16924
 |
Component:158; Jaccard:0.1903
 |
Component:222; Jaccard:0.20641
 |
Component:222; Jaccard:0.21735
 |
Correlation:0.54509
  |
Correlation:0.54509
  |
Correlation:0.2827
  |
Correlation:0.54509
  |
Correlation:0.54509
  |
Correlation:0.40255
  |
Correlation:0.40255
  |
Component:203; Jaccard:0.10519
 |
Component:203; Jaccard:0.12531
 |
Component:158; Jaccard:0.14714
 |
Component:203; Jaccard:0.16762
 |
Component:222; Jaccard:0.18645
 |
Component:158; Jaccard:0.20623
 |
Component:158; Jaccard:0.21522
 |
Correlation:0.2827
  |
Correlation:0.2827
  |
Correlation:0.54509
  |
Correlation:0.2827
  |
Correlation:0.40255
  |
Correlation:0.54509
  |
Correlation:0.54509
  |
Component:193; Jaccard:0.10391
 |
Component:250; Jaccard:0.12246
 |
Component:222; Jaccard:0.13822
 |
Component:222; Jaccard:0.16289
 |
Component:203; Jaccard:0.18521
 |
Component:203; Jaccard:0.19832
 |
Component:203; Jaccard:0.20154
 |
Correlation:0.28532
  |
Correlation:0.33912
  |
Correlation:0.40255
  |
Correlation:0.40255
  |
Correlation:0.2827
  |
Correlation:0.2827
  |
Correlation:0.2827
  |
Component:129; Jaccard:0.10285
 |
Component:193; Jaccard:0.11661
 |
Component:250; Jaccard:0.13658
 |
Component:250; Jaccard:0.14993
 |
Component:250; Jaccard:0.15907
 |
Component:026; Jaccard:0.16969
 |
Component:026; Jaccard:0.18223
 |
Correlation:0.48332
  |
Correlation:0.28532
  |
Correlation:0.33912
  |
Correlation:0.33912
  |
Correlation:0.33912
  |
Correlation:0.63831
  |
Correlation:0.63831
  |
Component:258; Jaccard:0.10049
 |
Component:222; Jaccard:0.11496
 |
Component:193; Jaccard:0.13068
 |
Component:193; Jaccard:0.14236
 |
Component:193; Jaccard:0.15265
 |
Component:250; Jaccard:0.16536
 |
Component:250; Jaccard:0.16298
 |
Correlation:0.41302
  |
Correlation:0.40255
  |
Correlation:0.28532
  |
Correlation:0.28532
  |
Correlation:0.28532
  |
Correlation:0.33912
  |
Correlation:0.33912
  |
Component:144; Jaccard:0.10029
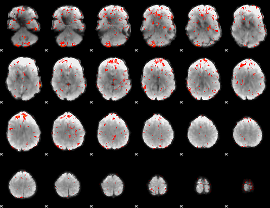 |
Component:144; Jaccard:0.11287
 |
Component:144; Jaccard:0.12562
 |
Component:144; Jaccard:0.13773
 |
Component:026; Jaccard:0.15024
 |
Component:193; Jaccard:0.1566
 |
Component:193; Jaccard:0.15506
 |
Correlation:0.053266
  |
Correlation:0.053266
  |
Correlation:0.053266
  |
Correlation:0.053266
  |
Correlation:0.63831
  |
Correlation:0.28532
  |
Correlation:0.28532
  |
Component:089; Jaccard:0.097507
 |
Component:258; Jaccard:0.11222
 |
Component:258; Jaccard:0.12372
 |
Component:258; Jaccard:0.13415
 |
Component:144; Jaccard:0.1466
 |
Component:144; Jaccard:0.15036
 |
Component:181; Jaccard:0.15494
 |
Correlation:0.25272
  |
Correlation:0.41302
  |
Correlation:0.41302
  |
Correlation:0.41302
  |
Correlation:0.053266
  |
Correlation:0.053266
  |
Correlation:0.28823
  |
Component:222; Jaccard:0.096656
 |
Component:089; Jaccard:0.10486
 |
Component:087; Jaccard:0.11047
 |
Component:026; Jaccard:0.12991
 |
Component:258; Jaccard:0.14151
 |
Component:087; Jaccard:0.14798
 |
Component:087; Jaccard:0.15321
 |
Correlation:0.40255
  |
Correlation:0.25272
  |
Correlation:0.34602
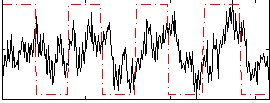  |
Correlation:0.63831
  |
Correlation:0.41302
  |
Correlation:0.34602
  |
Correlation:0.34602
  |
Cope: 05:"neg_MATH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
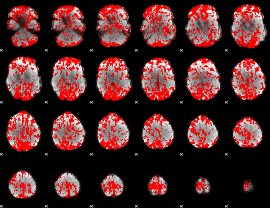 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
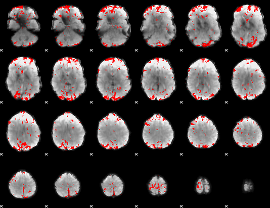 |
GLM binary result thresholded by 3.5
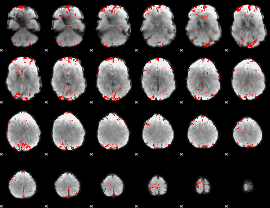 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
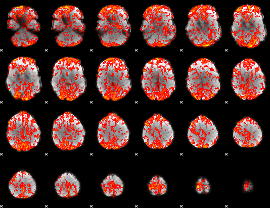 |
GLM Z-score result thresholded by 1.5
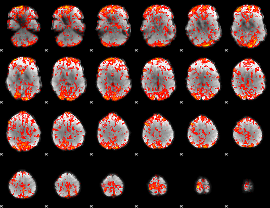 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:387; Jaccard:0.11549
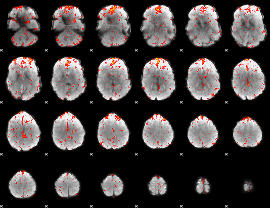 |
Component:387; Jaccard:0.13416
 |
Component:387; Jaccard:0.1476
 |
Component:387; Jaccard:0.15279
 |
Component:221; Jaccard:0.15681
 |
Component:221; Jaccard:0.14614
 |
Component:221; Jaccard:0.12011
 |
Correlation:-0.010528
  |
Correlation:-0.010528
  |
Correlation:-0.010528
  |
Correlation:-0.010528
  |
Correlation:0.31345
  |
Correlation:0.31345
  |
Correlation:0.31345
  |
Component:195; Jaccard:0.10832
 |
Component:368; Jaccard:0.1221
 |
Component:368; Jaccard:0.13896
 |
Component:221; Jaccard:0.15033
 |
Component:387; Jaccard:0.14189
 |
Component:368; Jaccard:0.12522
 |
Component:368; Jaccard:0.097607
 |
Correlation:-0.025999
  |
Correlation:0.18147
  |
Correlation:0.18147
  |
Correlation:0.31345
  |
Correlation:-0.010528
  |
Correlation:0.18147
  |
Correlation:0.18147
  |
Component:368; Jaccard:0.10447
 |
Component:195; Jaccard:0.11523
 |
Component:221; Jaccard:0.13077
 |
Component:368; Jaccard:0.14848
 |
Component:368; Jaccard:0.14114
 |
Component:387; Jaccard:0.11995
 |
Component:387; Jaccard:0.091953
 |
Correlation:0.18147
  |
Correlation:-0.025999
  |
Correlation:0.31345
  |
Correlation:0.18147
  |
Correlation:0.18147
  |
Correlation:-0.010528
  |
Correlation:-0.010528
  |
Component:275; Jaccard:0.098994
 |
Component:275; Jaccard:0.11003
 |
Component:275; Jaccard:0.11675
 |
Component:151; Jaccard:0.11819
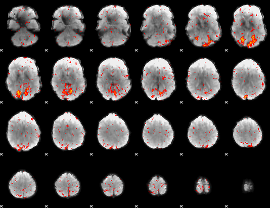 |
Component:001; Jaccard:0.11964
 |
Component:001; Jaccard:0.10974
 |
Component:001; Jaccard:0.089878
 |
Correlation:-0.11656
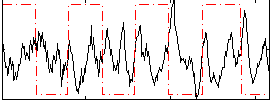  |
Correlation:-0.11656
  |
Correlation:-0.11656
  |
Correlation:0.10526
  |
Correlation:0.16677
  |
Correlation:0.16677
  |
Correlation:0.16677
  |
Component:340; Jaccard:0.087983
 |
Component:221; Jaccard:0.10757
 |
Component:195; Jaccard:0.11647
 |
Component:001; Jaccard:0.11595
 |
Component:151; Jaccard:0.11964
 |
Component:151; Jaccard:0.10948
 |
Component:151; Jaccard:0.087143
 |
Correlation:-0.31322
  |
Correlation:0.31345
  |
Correlation:-0.025999
  |
Correlation:0.16677
  |
Correlation:0.10526
  |
Correlation:0.10526
  |
Correlation:0.10526
  |
Component:355; Jaccard:0.08797
 |
Component:340; Jaccard:0.098439
 |
Component:151; Jaccard:0.10759
 |
Component:275; Jaccard:0.11489
 |
Component:340; Jaccard:0.10589
 |
Component:223; Jaccard:0.088995
 |
Component:223; Jaccard:0.070041
 |
Correlation:0.067372
 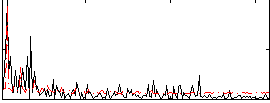 |
Correlation:-0.31322
  |
Correlation:0.10526
  |
Correlation:-0.11656
  |
Correlation:-0.31322
  |
Correlation:0.25823
  |
Correlation:0.25823
  |
Component:350; Jaccard:0.087726
 |
Component:355; Jaccard:0.098063
 |
Component:340; Jaccard:0.10737
 |
Component:340; Jaccard:0.11003
 |
Component:223; Jaccard:0.10337
 |
Component:340; Jaccard:0.088905
 |
Component:119; Jaccard:0.068778
 |
Correlation:-0.36729
  |
Correlation:0.067372
  |
Correlation:-0.31322
  |
Correlation:-0.31322
  |
Correlation:0.25823
  |
Correlation:-0.31322
  |
Correlation:0.11606
 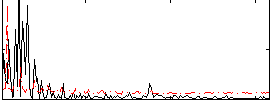 |
Component:221; Jaccard:0.08629
 |
Component:223; Jaccard:0.092485
 |
Component:355; Jaccard:0.10431
 |
Component:195; Jaccard:0.10934
 |
Component:275; Jaccard:0.10237
 |
Component:119; Jaccard:0.085423
 |
Component:340; Jaccard:0.06835
 |
Correlation:0.31345
  |
Correlation:0.25823
  |
Correlation:0.067372
  |
Correlation:-0.025999
  |
Correlation:-0.11656
  |
Correlation:0.11606
  |
Correlation:-0.31322
  |
Component:016; Jaccard:0.08463
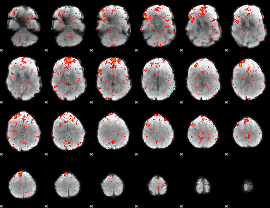 |
Component:151; Jaccard:0.092365
 |
Component:001; Jaccard:0.10331
 |
Component:223; Jaccard:0.10613
 |
Component:302; Jaccard:0.099046
 |
Component:302; Jaccard:0.084852
 |
Component:087; Jaccard:0.062702
 |
Correlation:-0.07744
  |
Correlation:0.10526
  |
Correlation:0.16677
  |
Correlation:0.25823
  |
Correlation:0.36126
  |
Correlation:0.36126
  |
Correlation:0.33556
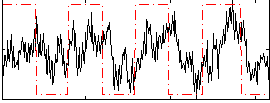  |
Component:180; Jaccard:0.080138
 |
Component:016; Jaccard:0.091068
 |
Component:223; Jaccard:0.10151
 |
Component:302; Jaccard:0.10399
 |
Component:195; Jaccard:0.094356
 |
Component:087; Jaccard:0.079133
 |
Component:033; Jaccard:0.06146
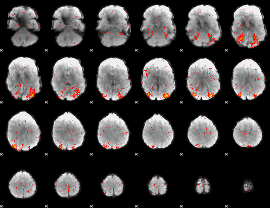 |
Correlation:0.10165
  |
Correlation:-0.07744
  |
Correlation:0.25823
  |
Correlation:0.36126
  |
Correlation:-0.025999
  |
Correlation:0.33556
  |
Correlation:0.34662
 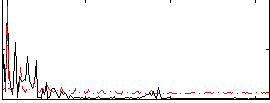 |
Cope: 06:"neg_STORY" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:350; Jaccard:0.11901
 |
Component:340; Jaccard:0.14517
 |
Component:340; Jaccard:0.17472
 |
Component:340; Jaccard:0.19937
 |
Component:340; Jaccard:0.21338
 |
Component:340; Jaccard:0.21095
 |
Component:340; Jaccard:0.18845
 |
Correlation:0.39044
  |
Correlation:0.31908
  |
Correlation:0.31908
  |
Correlation:0.31908
  |
Correlation:0.31908
  |
Correlation:0.31908
  |
Correlation:0.31908
  |
Component:340; Jaccard:0.11895
 |
Component:350; Jaccard:0.13649
 |
Component:350; Jaccard:0.15273
 |
Component:350; Jaccard:0.15859
 |
Component:350; Jaccard:0.15478
 |
Component:306; Jaccard:0.13889
 |
Component:306; Jaccard:0.11624
 |
Correlation:0.31908
  |
Correlation:0.39044
  |
Correlation:0.39044
  |
Correlation:0.39044
  |
Correlation:0.39044
  |
Correlation:0.5956
  |
Correlation:0.5956
  |
Component:109; Jaccard:0.1181
 |
Component:109; Jaccard:0.13576
 |
Component:109; Jaccard:0.14971
 |
Component:109; Jaccard:0.15386
 |
Component:306; Jaccard:0.15089
 |
Component:350; Jaccard:0.13552
 |
Component:133; Jaccard:0.11406
 |
Correlation:0.33462
  |
Correlation:0.33462
  |
Correlation:0.33462
  |
Correlation:0.33462
  |
Correlation:0.5956
  |
Correlation:0.39044
  |
Correlation:0.32422
  |
Component:195; Jaccard:0.10759
 |
Component:306; Jaccard:0.12222
 |
Component:306; Jaccard:0.14074
 |
Component:306; Jaccard:0.15322
 |
Component:109; Jaccard:0.14699
 |
Component:133; Jaccard:0.13277
 |
Component:350; Jaccard:0.11155
 |
Correlation:0.027689
  |
Correlation:0.5956
  |
Correlation:0.5956
  |
Correlation:0.5956
  |
Correlation:0.33462
  |
Correlation:0.32422
  |
Correlation:0.39044
  |
Component:368; Jaccard:0.10437
 |
Component:368; Jaccard:0.11921
 |
Component:368; Jaccard:0.12968
 |
Component:370; Jaccard:0.14006
 |
Component:370; Jaccard:0.13875
 |
Component:370; Jaccard:0.12956
 |
Component:370; Jaccard:0.10761
 |
Correlation:-0.15597
  |
Correlation:-0.15597
  |
Correlation:-0.15597
  |
Correlation:0.52878
  |
Correlation:0.52878
  |
Correlation:0.52878
  |
Correlation:0.52878
  |
Component:306; Jaccard:0.10295
 |
Component:326; Jaccard:0.1136
 |
Component:370; Jaccard:0.12905
 |
Component:368; Jaccard:0.1369
 |
Component:133; Jaccard:0.13566
 |
Component:109; Jaccard:0.12765
 |
Component:368; Jaccard:0.10394
 |
Correlation:0.5956
  |
Correlation:0.22525
  |
Correlation:0.52878
  |
Correlation:-0.15597
  |
Correlation:0.32422
  |
Correlation:0.33462
  |
Correlation:-0.15597
  |
Component:326; Jaccard:0.10277
 |
Component:370; Jaccard:0.11176
 |
Component:263; Jaccard:0.12169
 |
Component:133; Jaccard:0.13173
 |
Component:368; Jaccard:0.13463
 |
Component:368; Jaccard:0.12262
 |
Component:109; Jaccard:0.1021
 |
Correlation:0.22525
  |
Correlation:0.52878
  |
Correlation:0.4822
  |
Correlation:0.32422
  |
Correlation:-0.15597
  |
Correlation:-0.15597
  |
Correlation:0.33462
  |
Component:387; Jaccard:0.10239
 |
Component:195; Jaccard:0.11127
 |
Component:326; Jaccard:0.11967
 |
Component:263; Jaccard:0.12802
 |
Component:263; Jaccard:0.12491
 |
Component:263; Jaccard:0.11344
 |
Component:001; Jaccard:0.095814
 |
Correlation:0.015847
  |
Correlation:0.027689
  |
Correlation:0.22525
  |
Correlation:0.4822
  |
Correlation:0.4822
  |
Correlation:0.4822
  |
Correlation:-0.19204
  |
Component:218; Jaccard:0.095756
 |
Component:387; Jaccard:0.11117
 |
Component:133; Jaccard:0.11947
 |
Component:218; Jaccard:0.12411
 |
Component:183; Jaccard:0.12237
 |
Component:183; Jaccard:0.11085
 |
Component:183; Jaccard:0.094108
 |
Correlation:0.55101
  |
Correlation:0.015847
  |
Correlation:0.32422
  |
Correlation:0.55101
  |
Correlation:0.45356
  |
Correlation:0.45356
  |
Correlation:0.45356
  |
Component:263; Jaccard:0.09556
 |
Component:263; Jaccard:0.10836
 |
Component:218; Jaccard:0.11899
 |
Component:183; Jaccard:0.12329
 |
Component:218; Jaccard:0.11913
 |
Component:001; Jaccard:0.10846
 |
Component:263; Jaccard:0.091239
 |
Correlation:0.4822
  |
Correlation:0.4822
  |
Correlation:0.55101
  |
Correlation:0.45356
  |
Correlation:0.55101
  |
Correlation:-0.19204
  |
Correlation:0.4822
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































