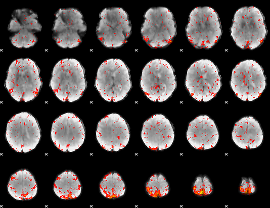
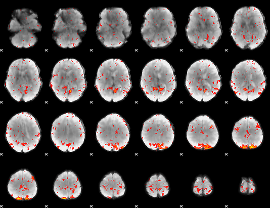
Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"MATCH" BACK
|
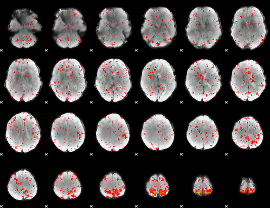
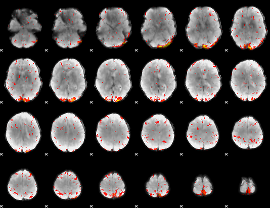
GLM result group-wise
 |
GLM binary result thresholded by 1
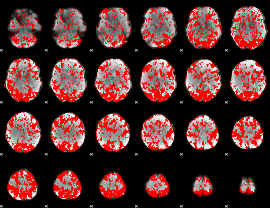 |
GLM binary result thresholded by 1.5
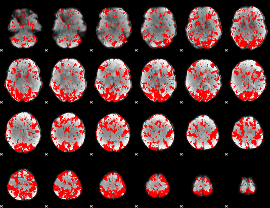 |
GLM binary result thresholded by 2
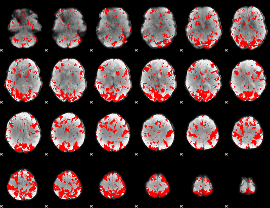 |
GLM binary result thresholded by 2.5
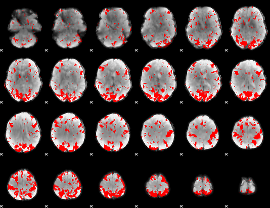 |
GLM binary result thresholded by 3
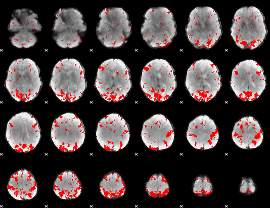 |
GLM binary result thresholded by 3.5
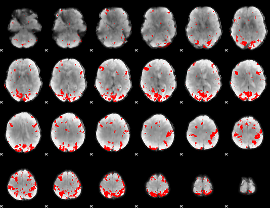 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
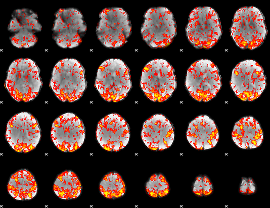 |
GLM Z-score result thresholded by 2
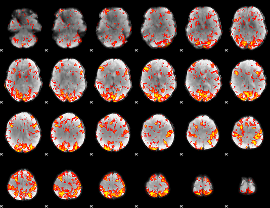 |
GLM Z-score result thresholded by 2.5
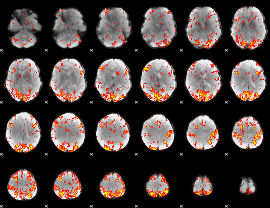 |
GLM Z-score result thresholded by 3
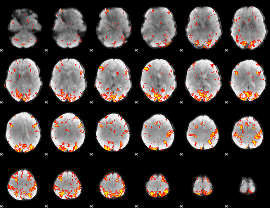 |
GLM Z-score result thresholded by 3.5
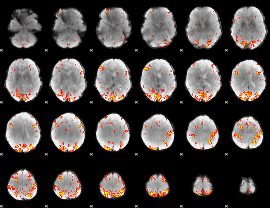 |
GLM Z-score result thresholded by 4
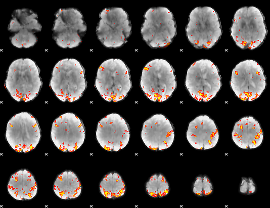 |
Component:217; Jaccard:0.12583
 |
Component:217; Jaccard:0.16387
 |
Component:217; Jaccard:0.21174
 |
Component:217; Jaccard:0.26401
 |
Component:217; Jaccard:0.30769
 |
Component:217; Jaccard:0.32609
 |
Component:217; Jaccard:0.31859
 |
Correlation:0.18936
  |
Correlation:0.18936
  |
Correlation:0.18936
  |
Correlation:0.18936
  |
Correlation:0.18936
  |
Correlation:0.18936
  |
Correlation:0.18936
  |
Component:323; Jaccard:0.11572
 |
Component:128; Jaccard:0.13814
 |
Component:128; Jaccard:0.17175
 |
Component:128; Jaccard:0.20494
 |
Component:128; Jaccard:0.23192
 |
Component:128; Jaccard:0.24759
 |
Component:128; Jaccard:0.24938
 |
Correlation:0.028116
 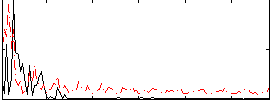 |
Correlation:0.3007
  |
Correlation:0.3007
  |
Correlation:0.3007
  |
Correlation:0.3007
  |
Correlation:0.3007
  |
Correlation:0.3007
  |
Component:128; Jaccard:0.11019
 |
Component:323; Jaccard:0.13397
 |
Component:069; Jaccard:0.16705
 |
Component:069; Jaccard:0.1997
 |
Component:069; Jaccard:0.22895
 |
Component:069; Jaccard:0.24345
 |
Component:069; Jaccard:0.23978
 |
Correlation:0.3007
  |
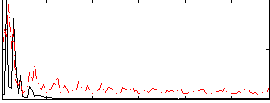
Correlation:0.028116
  |
Correlation:0.1173
  |
Correlation:0.1173
  |
Correlation:0.1173
  |
Correlation:0.1173
  |
Correlation:0.1173
  |
Component:007; Jaccard:0.10721
 |
Component:069; Jaccard:0.13268
 |
Component:323; Jaccard:0.15317
 |
Component:297; Jaccard:0.17622
 |
Component:297; Jaccard:0.19151
 |
Component:297; Jaccard:0.1986
 |
Component:297; Jaccard:0.19285
 |
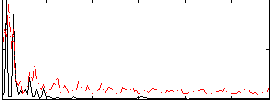
Correlation:0.017691
  |
Correlation:0.1173
  |
Correlation:0.028116
  |
Correlation:0.2005
  |
Correlation:0.2005
  |
Correlation:0.2005
  |
Correlation:0.2005
  |
Component:069; Jaccard:0.10476
 |
Component:297; Jaccard:0.12574
 |
Component:297; Jaccard:0.1515
 |
Component:323; Jaccard:0.1694
 |
Component:323; Jaccard:0.17635
 |
Component:323; Jaccard:0.17002
 |
Component:323; Jaccard:0.1585
 |
Correlation:0.1173
  |
Correlation:0.2005
  |
Correlation:0.2005
  |
Correlation:0.028116
  |
Correlation:0.028116
  |
Correlation:0.028116
  |
Correlation:0.028116
  |
Component:297; Jaccard:0.10278
 |
Component:007; Jaccard:0.11717
 |
Component:043; Jaccard:0.12477
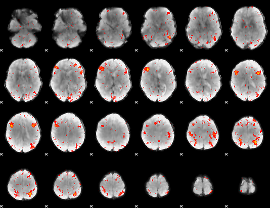 |
Component:043; Jaccard:0.1439
 |
Component:043; Jaccard:0.15706
 |
Component:043; Jaccard:0.15856
 |
Component:043; Jaccard:0.15393
 |
Correlation:0.2005
  |
Correlation:0.017691
  |
Correlation:0.14597
  |
Correlation:0.14597
  |
Correlation:0.14597
  |
Correlation:0.14597
  |
Correlation:0.14597
  |
Component:364; Jaccard:0.10033
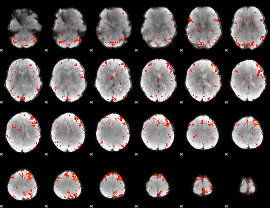 |
Component:364; Jaccard:0.10997
 |
Component:007; Jaccard:0.1245
 |
Component:387; Jaccard:0.13862
 |
Component:321; Jaccard:0.14725
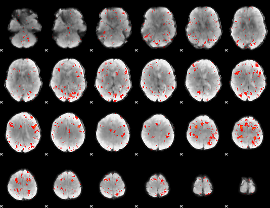 |
Component:387; Jaccard:0.14616
 |
Component:387; Jaccard:0.13682
 |
Correlation:0.069227
  |
Correlation:0.069227
  |
Correlation:0.017691
  |
Correlation:0.054399
 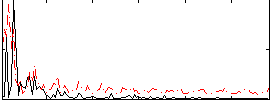 |
Correlation:0.20306
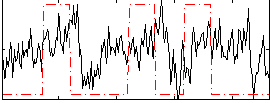  |
Correlation:0.054399
  |
Correlation:0.054399
  |
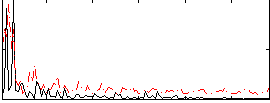
Component:228; Jaccard:0.099426
 |
Component:228; Jaccard:0.10917
 |
Component:376; Jaccard:0.12322
 |
Component:321; Jaccard:0.13676
 |
Component:387; Jaccard:0.14653
 |
Component:321; Jaccard:0.14379
 |
Component:321; Jaccard:0.13673
 |
Correlation:0.18982
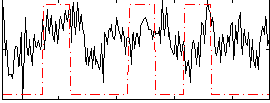 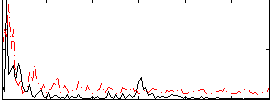 |
Correlation:0.18982
  |
Correlation:0.2503
 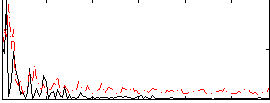 |
Correlation:0.20306
  |
Correlation:0.054399
  |
Correlation:0.20306
  |
Correlation:0.20306
  |
Component:376; Jaccard:0.092968
 |
Component:376; Jaccard:0.10855
 |
Component:387; Jaccard:0.12271
 |
Component:376; Jaccard:0.13263
 |
Component:376; Jaccard:0.13611
 |
Component:376; Jaccard:0.12842
 |
Component:376; Jaccard:0.11589
 |
Correlation:0.2503
  |
Correlation:0.2503
  |
Correlation:0.054399
  |
Correlation:0.2503
  |
Correlation:0.2503
  |
Correlation:0.2503
  |
Correlation:0.2503
  |
Component:255; Jaccard:0.09278
 |
Component:255; Jaccard:0.10616
 |
Component:321; Jaccard:0.12129
 |
Component:007; Jaccard:0.12632
 |
Component:012; Jaccard:0.12635
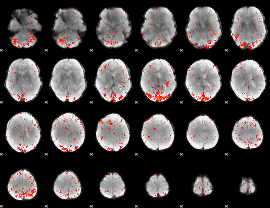 |
Component:012; Jaccard:0.12052
 |
Component:205; Jaccard:0.11184
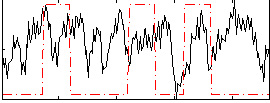
 |
Correlation:0.045543
 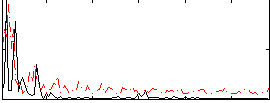 |
Correlation:0.045543
  |
Correlation:0.20306
  |
Correlation:0.017691
  |
Correlation:0.08899
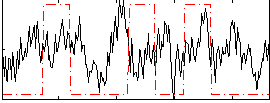 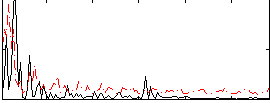 |
Correlation:0.08899
  |
Correlation:0.037864
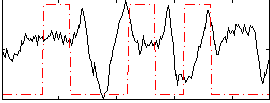 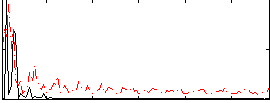 |
Cope: 02:"REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
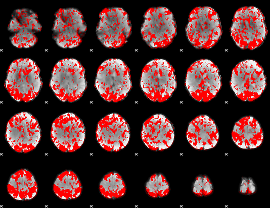 |
GLM binary result thresholded by 1.5
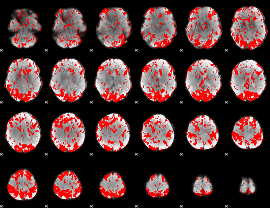 |
GLM binary result thresholded by 2
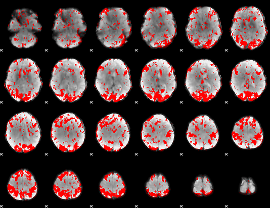 |
GLM binary result thresholded by 2.5
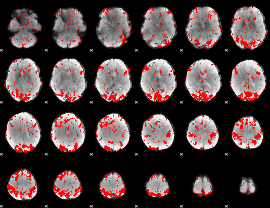 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
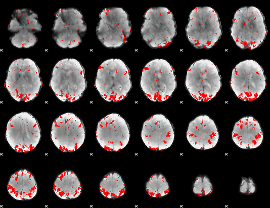 |
GLM binary result thresholded by 4
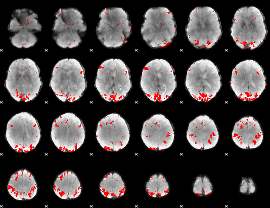 |
GLM Z-score result thresholded by 1
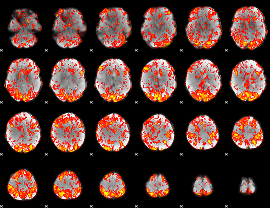 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
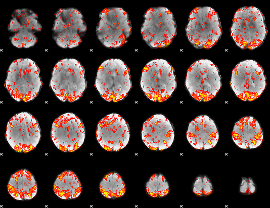 |
GLM Z-score result thresholded by 2.5
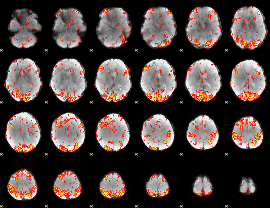 |
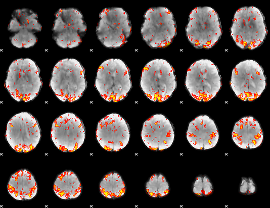
GLM Z-score result thresholded by 3
 |
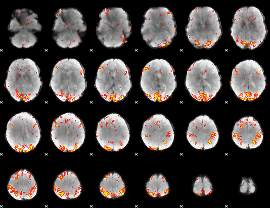
GLM Z-score result thresholded by 3.5
 |
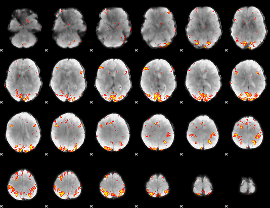
GLM Z-score result thresholded by 4
 |
Component:217; Jaccard:0.12555
 |
Component:217; Jaccard:0.1565
 |
Component:217; Jaccard:0.1914
 |
Component:217; Jaccard:0.22981
 |
Component:217; Jaccard:0.26136
 |
Component:217; Jaccard:0.27766
 |
Component:217; Jaccard:0.27835
 |
Correlation:0.059971
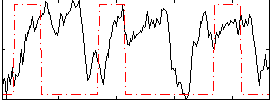  |
Correlation:0.059971
  |
Correlation:0.059971
  |
Correlation:0.059971
  |
Correlation:0.059971
  |
Correlation:0.059971
  |
Correlation:0.059971
  |
Component:128; Jaccard:0.10901
 |
Component:128; Jaccard:0.13164
 |
Component:128; Jaccard:0.15775
 |
Component:128; Jaccard:0.18658
 |
Component:128; Jaccard:0.2123
 |
Component:128; Jaccard:0.23185
 |
Component:128; Jaccard:0.23645
 |
Correlation:0.0073984
  |
Correlation:0.0073984
  |
Correlation:0.0073984
  |
Correlation:0.0073984
  |
Correlation:0.0073984
  |
Correlation:0.0073984
  |
Correlation:0.0073984
  |
Component:251; Jaccard:0.10774
 |
Component:069; Jaccard:0.12651
 |
Component:069; Jaccard:0.151
 |
Component:069; Jaccard:0.17773
 |
Component:069; Jaccard:0.19959
 |
Component:069; Jaccard:0.21353
 |
Component:069; Jaccard:0.21843
 |
Correlation:0.1171
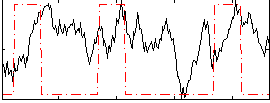 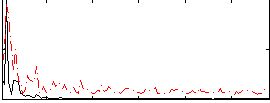 |
Correlation:0.16414
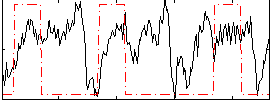 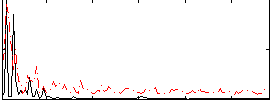 |
Correlation:0.16414
  |
Correlation:0.16414
  |
Correlation:0.16414
  |
Correlation:0.16414
  |
Correlation:0.16414
  |
Component:231; Jaccard:0.10421
 |
Component:251; Jaccard:0.12304
 |
Component:251; Jaccard:0.1374
 |
Component:297; Jaccard:0.15095
 |
Component:297; Jaccard:0.16159
 |
Component:297; Jaccard:0.16614
 |
Component:297; Jaccard:0.16545
 |
Correlation:0.28019
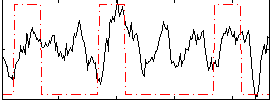 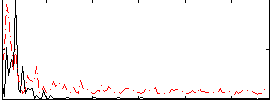 |
Correlation:0.1171
  |
Correlation:0.1171
  |
Correlation:0.24983
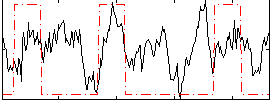 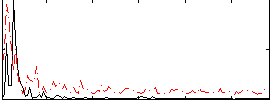 |
Correlation:0.24983
  |
Correlation:0.24983
  |
Correlation:0.24983
  |
Component:069; Jaccard:0.10403
 |
Component:255; Jaccard:0.12051
 |
Component:255; Jaccard:0.13544
 |
Component:251; Jaccard:0.14699
 |
Component:255; Jaccard:0.14844
 |
Component:205; Jaccard:0.14719
 |
Component:043; Jaccard:0.14176
 |
Correlation:0.16414
  |
Correlation:0.12874
  |
Correlation:0.12874
  |
Correlation:0.1171
  |
Correlation:0.12874
  |
Correlation:-0.025861
  |
Correlation:0.056398
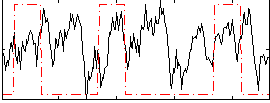 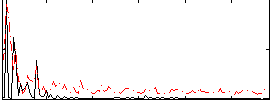 |
Component:255; Jaccard:0.10392
 |
Component:231; Jaccard:0.1166
 |
Component:297; Jaccard:0.13385
 |
Component:255; Jaccard:0.14529
 |
Component:205; Jaccard:0.14698
 |
Component:043; Jaccard:0.14324
 |
Component:205; Jaccard:0.14166
 |
Correlation:0.12874
  |
Correlation:0.28019
  |
Correlation:0.24983
  |
Correlation:0.12874
  |
Correlation:-0.025861
  |
Correlation:0.056398
  |
Correlation:-0.025861
  |
Component:323; Jaccard:0.099391
 |
Component:297; Jaccard:0.11594
 |
Component:231; Jaccard:0.12676
 |
Component:205; Jaccard:0.13931
 |
Component:251; Jaccard:0.14637
 |
Component:255; Jaccard:0.14147
 |
Component:387; Jaccard:0.13508
 |
Correlation:0.15749
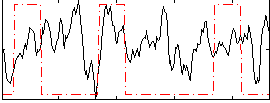 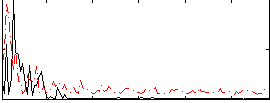 |
Correlation:0.24983
  |
Correlation:0.28019
  |
Correlation:-0.025861
  |
Correlation:0.1171
  |
Correlation:0.12874
  |
Correlation:0.35072
  |
Component:297; Jaccard:0.098938
 |
Component:205; Jaccard:0.1128
 |
Component:205; Jaccard:0.1266
 |
Component:231; Jaccard:0.13403
 |
Component:043; Jaccard:0.13961
 |
Component:387; Jaccard:0.14017
 |
Component:255; Jaccard:0.12777
 |
Correlation:0.24983
  |
Correlation:-0.025861
  |
Correlation:-0.025861
  |
Correlation:0.28019
  |
Correlation:0.056398
  |
Correlation:0.35072
  |
Correlation:0.12874
  |
Component:012; Jaccard:0.096734
 |
Component:323; Jaccard:0.10948
 |
Component:323; Jaccard:0.11835
 |
Component:387; Jaccard:0.13018
 |
Component:387; Jaccard:0.13721
 |
Component:251; Jaccard:0.13676
 |
Component:012; Jaccard:0.12323
 |
Correlation:0.14984
  |
Correlation:0.15749
  |
Correlation:0.15749
  |
Correlation:0.35072
  |
Correlation:0.35072
  |
Correlation:0.1171
  |
Correlation:0.14984
  |
Component:205; Jaccard:0.096522
 |
Component:012; Jaccard:0.1082
 |
Component:012; Jaccard:0.1182
 |
Component:043; Jaccard:0.12883
 |
Component:231; Jaccard:0.13674
 |
Component:231; Jaccard:0.1336
 |
Component:231; Jaccard:0.12314
 |
Correlation:-0.025861
  |
Correlation:0.14984
  |
Correlation:0.14984
  |
Correlation:0.056398
  |
Correlation:0.28019
  |
Correlation:0.28019
  |
Correlation:0.28019
  |
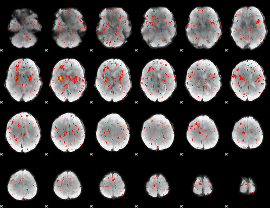
Cope: 03:"MATCH-REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
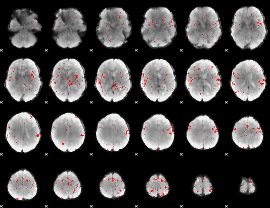
GLM binary result thresholded by 3
 |
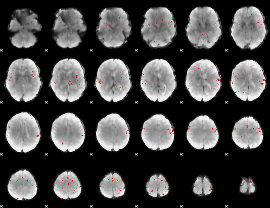
GLM binary result thresholded by 3.5
 |
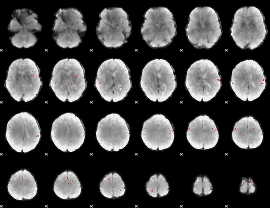
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
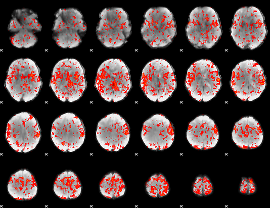
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
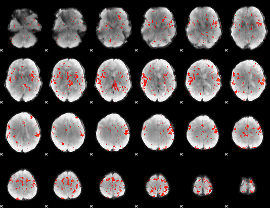 |
GLM Z-score result thresholded by 3
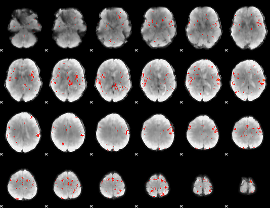 |
GLM Z-score result thresholded by 3.5
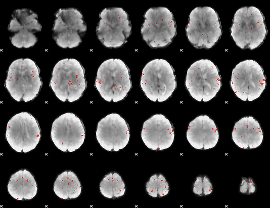 |
GLM Z-score result thresholded by 4
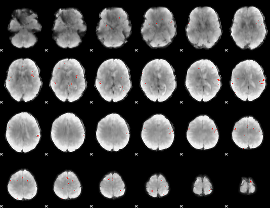 |
Component:376; Jaccard:0.10088
 |
Component:376; Jaccard:0.11748
 |
Component:376; Jaccard:0.12704
 |
Component:376; Jaccard:0.11362
 |
Component:376; Jaccard:0.076948
 |
Component:376; Jaccard:0.039118
 |
Component:376; Jaccard:0.01497
 |
Correlation:0.19005
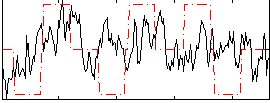 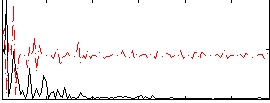 |
Correlation:0.19005
  |
Correlation:0.19005
  |
Correlation:0.19005
  |
Correlation:0.19005
  |
Correlation:0.19005
  |
Correlation:0.19005
  |
Component:354; Jaccard:0.098146
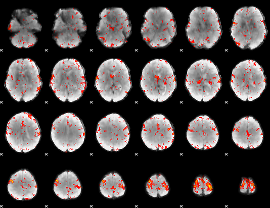 |
Component:228; Jaccard:0.10091
 |
Component:210; Jaccard:0.098764
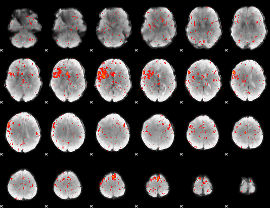 |
Component:210; Jaccard:0.086379
 |
Component:340; Jaccard:0.05674
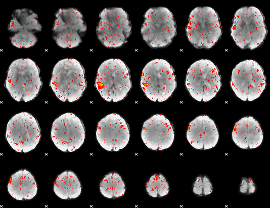 |
Component:340; Jaccard:0.030452
 |
Component:340; Jaccard:0.011757
 |
Correlation:0.17806
  |
Correlation:0.19637
  |
Correlation:0.33406
  |
Correlation:0.33406
  |
Correlation:0.28002
  |
Correlation:0.28002
  |
Correlation:0.28002
  |
Component:228; Jaccard:0.096147
 |
Component:378; Jaccard:0.09827
 |
Component:374; Jaccard:0.097046
 |
Component:183; Jaccard:0.084124
 |
Component:183; Jaccard:0.056325
 |
Component:183; Jaccard:0.028116
 |
Component:186; Jaccard:0.010273
 |
Correlation:0.19637
  |
Correlation:0.096681
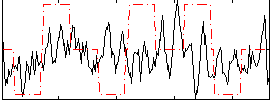  |
Correlation:0.020012
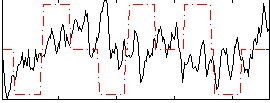 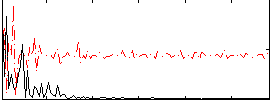 |
Correlation:0.17449
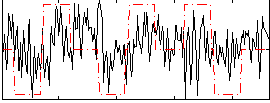 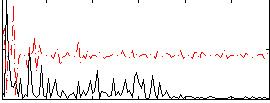 |
Correlation:0.17449
  |
Correlation:0.17449
  |
Correlation:0.26371
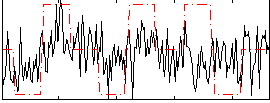  |
Component:378; Jaccard:0.091923
 |
Component:340; Jaccard:0.096666
 |
Component:340; Jaccard:0.09409
 |
Component:340; Jaccard:0.081965
 |
Component:210; Jaccard:0.05373
 |
Component:210; Jaccard:0.027624
 |
Component:374; Jaccard:0.010093
 |
Correlation:0.096681
  |
Correlation:0.28002
  |
Correlation:0.28002
  |
Correlation:0.28002
  |
Correlation:0.33406
  |
Correlation:0.33406
  |
Correlation:0.020012
  |
Component:340; Jaccard:0.08898
 |
Component:374; Jaccard:0.096569
 |
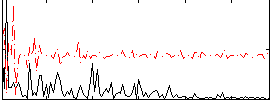
Component:006; Jaccard:0.093526
 |
Component:374; Jaccard:0.07797
 |
Component:374; Jaccard:0.050441
 |
Component:374; Jaccard:0.026799
 |
Component:256; Jaccard:0.009979
 |
Correlation:0.28002
  |
Correlation:0.020012
  |
Correlation:-0.0057267
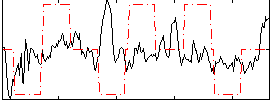  |
Correlation:0.020012
  |
Correlation:0.020012
  |
Correlation:0.020012
  |
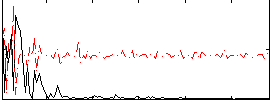
Correlation:0.12643
  |
Component:362; Jaccard:0.088527
 |
Component:354; Jaccard:0.09611
 |
Component:378; Jaccard:0.093329
 |
Component:006; Jaccard:0.075209
 |
Component:186; Jaccard:0.046955
 |
Component:186; Jaccard:0.024839
 |
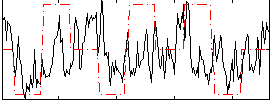
Component:089; Jaccard:0.009764
 |
Correlation:0.20978
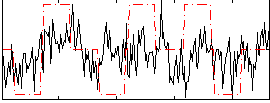 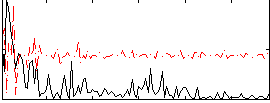 |
Correlation:0.17806
  |
Correlation:0.096681
  |
Correlation:-0.0057267
  |
Correlation:0.26371
  |
Correlation:0.26371
  |
Correlation:0.34046
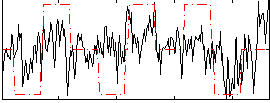 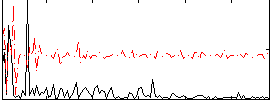 |
Component:006; Jaccard:0.088332
 |
Component:006; Jaccard:0.095758
 |
Component:228; Jaccard:0.092056
 |
Component:173; Jaccard:0.075092
 |
Component:006; Jaccard:0.046636
 |
Component:256; Jaccard:0.023624
 |
Component:378; Jaccard:0.009512
 |
Correlation:-0.0057267
  |
Correlation:-0.0057267
  |
Correlation:0.19637
  |
Correlation:0.17103
  |
Correlation:-0.0057267
  |
Correlation:0.12643
  |
Correlation:0.096681
  |
Component:374; Jaccard:0.088276
 |
Component:210; Jaccard:0.09533
 |
Component:183; Jaccard:0.090553
 |
Component:378; Jaccard:0.073612
 |
Component:256; Jaccard:0.04561
 |
Component:006; Jaccard:0.02339
 |
Component:210; Jaccard:0.009349
 |
Correlation:0.020012
  |
Correlation:0.33406
  |
Correlation:0.17449
  |
Correlation:0.096681
  |
Correlation:0.12643
  |
Correlation:-0.0057267
  |
Correlation:0.33406
  |
Component:165; Jaccard:0.086203
 |
Component:362; Jaccard:0.092016
 |
Component:362; Jaccard:0.086448
 |
Component:229; Jaccard:0.07313
 |
Component:173; Jaccard:0.045055
 |
Component:362; Jaccard:0.023362
 |
Component:183; Jaccard:0.0091
 |
Correlation:0.087438
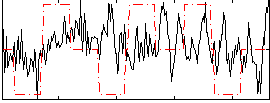 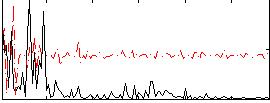 |
Correlation:0.20978
  |
Correlation:0.20978
  |
Correlation:0.20609
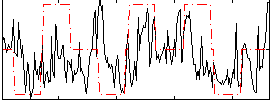 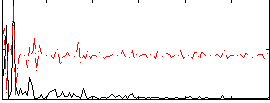 |
Correlation:0.17103
  |
Correlation:0.20978
  |
Correlation:0.17449
  |
Component:210; Jaccard:0.083169
 |
Component:165; Jaccard:0.08864
 |
Component:173; Jaccard:0.08517
 |
Component:186; Jaccard:0.070826
 |
Component:229; Jaccard:0.044726
 |
Component:378; Jaccard:0.023338
 |
Component:362; Jaccard:0.008975
 |
Correlation:0.33406
  |
Correlation:0.087438
  |
Correlation:0.17103
  |
Correlation:0.26371
  |
Correlation:0.20609
  |
Correlation:0.096681
  |
Correlation:0.20978
  |
Cope: 04:"REL-MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
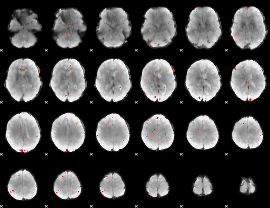 |
GLM binary result thresholded by 3.5
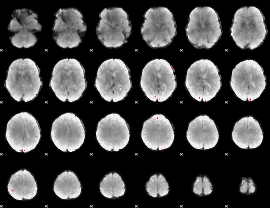 |
GLM binary result thresholded by 4
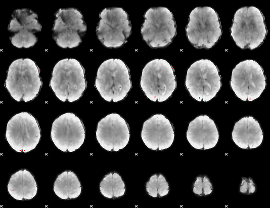 |
GLM Z-score result thresholded by 1
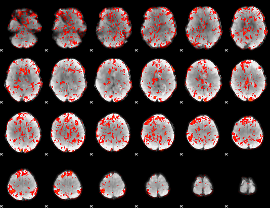 |
GLM Z-score result thresholded by 1.5
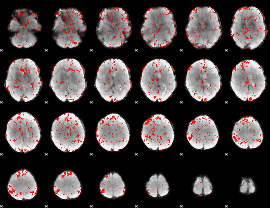 |
GLM Z-score result thresholded by 2
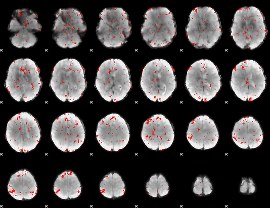 |
GLM Z-score result thresholded by 2.5
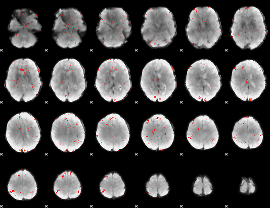 |
GLM Z-score result thresholded by 3
 |
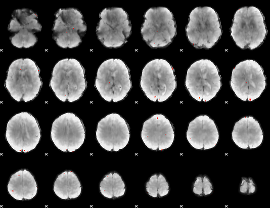
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
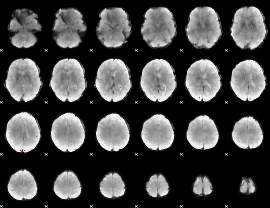 |
Component:231; Jaccard:0.095389
 |
Component:231; Jaccard:0.096267
 |
Component:137; Jaccard:0.087904
 |
Component:137; Jaccard:0.06342
 |
Component:137; Jaccard:0.032758
 |
Component:137; Jaccard:0.013444
 |
Component:137; Jaccard:0.005132
 |
Correlation:0.17797
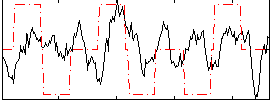 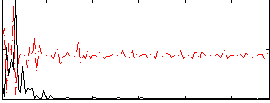 |
Correlation:0.17797
  |
Correlation:0.2421
 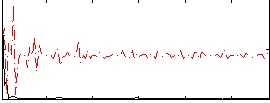 |
Correlation:0.2421
  |
Correlation:0.2421
  |
Correlation:0.2421
  |
Correlation:0.2421
  |
Component:251; Jaccard:0.088024
 |
Component:137; Jaccard:0.090076
 |
Component:231; Jaccard:0.082107
 |
Component:231; Jaccard:0.057759
 |
Component:231; Jaccard:0.0302
 |
Component:231; Jaccard:0.011503
 |
Component:231; Jaccard:0.004961
 |
Correlation:0.045813
 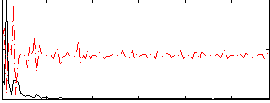 |
Correlation:0.2421
  |
Correlation:0.17797
  |
Correlation:0.17797
  |
Correlation:0.17797
  |
Correlation:0.17797
  |
Correlation:0.17797
  |
Component:023; Jaccard:0.086272
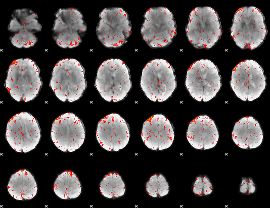 |
Component:319; Jaccard:0.084523
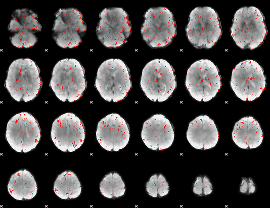 |
Component:319; Jaccard:0.070818
 |
Component:023; Jaccard:0.041122
 |
Component:246; Jaccard:0.017813
 |
Component:293; Jaccard:0.006143
 |
Component:293; Jaccard:0.003226
 |
Correlation:0.11891
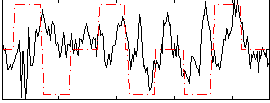 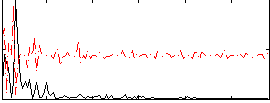 |
Correlation:0.24302
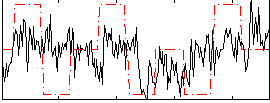 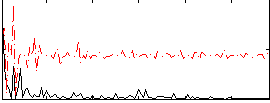 |
Correlation:0.24302
  |
Correlation:0.11891
  |
Correlation:0.27978
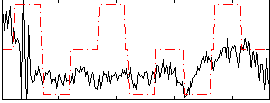  |
Correlation:0.25995
  |
Correlation:0.25995
  |
Component:053; Jaccard:0.083159
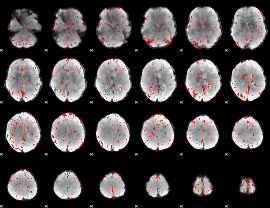 |
Component:251; Jaccard:0.083005
 |
Component:246; Jaccard:0.0676
 |
Component:319; Jaccard:0.040639
 |
Component:023; Jaccard:0.017023
 |
Component:351; Jaccard:0.005952
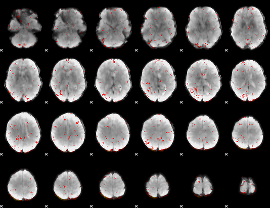 |
Component:031; Jaccard:0.002874
 |
Correlation:0.19258
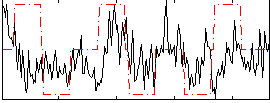 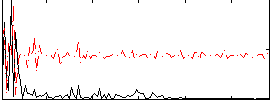 |
Correlation:0.045813
  |
Correlation:0.27978
  |
Correlation:0.24302
  |
Correlation:0.11891
  |
Correlation:0.18999
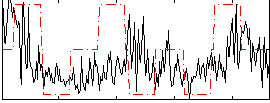  |
Correlation:0.034346
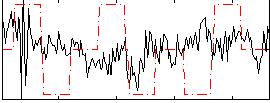 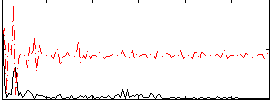 |
Component:319; Jaccard:0.082809
 |
Component:246; Jaccard:0.081463
 |
Component:023; Jaccard:0.067217
 |
Component:246; Jaccard:0.040072
 |
Component:293; Jaccard:0.016984
 |
Component:053; Jaccard:0.005883
 |
Component:387; Jaccard:0.002769
 |
Correlation:0.24302
  |
Correlation:0.27978
  |
Correlation:0.11891
  |
Correlation:0.27978
  |
Correlation:0.25995
  |
Correlation:0.19258
  |
Correlation:0.17563
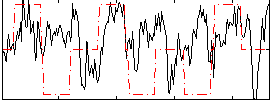 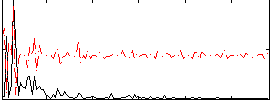 |
Component:137; Jaccard:0.079782
 |
Component:023; Jaccard:0.081379
 |
Component:369; Jaccard:0.062388
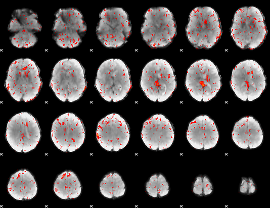 |
Component:369; Jaccard:0.036917
 |
Component:351; Jaccard:0.016788
 |
Component:023; Jaccard:0.005275
 |
Component:132; Jaccard:0.00246
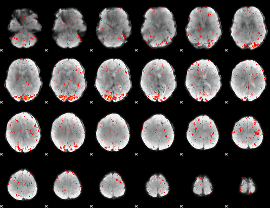 |
Correlation:0.2421
  |
Correlation:0.11891
  |
Correlation:0.20867
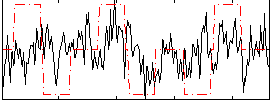 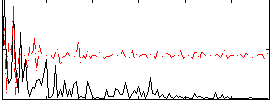 |
Correlation:0.20867
  |
Correlation:0.18999
  |
Correlation:0.11891
  |
Correlation:-0.040704
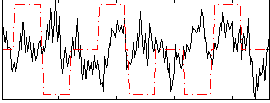  |
Component:246; Jaccard:0.078543
 |
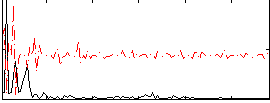
Component:283; Jaccard:0.077642
 |
Component:251; Jaccard:0.062011
 |
Component:351; Jaccard:0.035714
 |
Component:053; Jaccard:0.016143
 |
Component:031; Jaccard:0.005103
 |
Component:012; Jaccard:0.002422
 |
Correlation:0.27978
  |
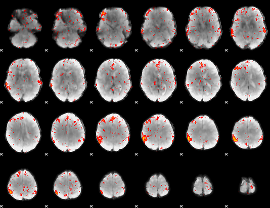
Correlation:0.045803
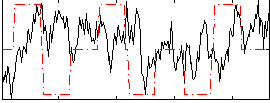 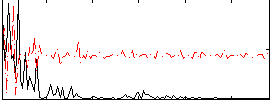 |
Correlation:0.045813
  |
Correlation:0.18999
  |
Correlation:0.19258
  |
Correlation:0.034346
  |
Correlation:0.036063
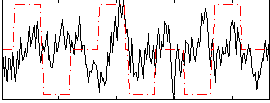 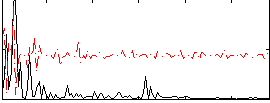 |
Component:283; Jaccard:0.076517
 |
Component:369; Jaccard:0.077306
 |
Component:371; Jaccard:0.060086
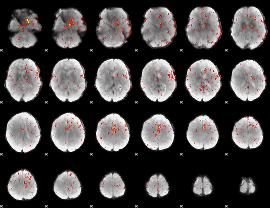 |
Component:283; Jaccard:0.035569
 |
Component:319; Jaccard:0.015743
 |
Component:246; Jaccard:0.004735
 |
Component:128; Jaccard:0.002049
 |
Correlation:0.045803
  |
Correlation:0.20867
  |
Correlation:0.10273
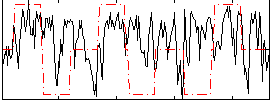  |
Correlation:0.045803
  |
Correlation:0.24302
  |
Correlation:0.27978
  |
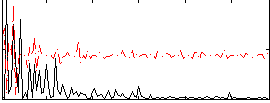
Correlation:-0.17384
  |
Component:369; Jaccard:0.076284
 |
Component:053; Jaccard:0.074974
 |
Component:283; Jaccard:0.059648
 |
Component:251; Jaccard:0.035534
 |
Component:369; Jaccard:0.015283
 |
Component:387; Jaccard:0.004668
 |
Component:351; Jaccard:0.0018
 |
Correlation:0.20867
  |
Correlation:0.19258
  |
Correlation:0.045803
  |
Correlation:0.045813
  |
Correlation:0.20867
  |
Correlation:0.17563
  |
Correlation:0.18999
  |
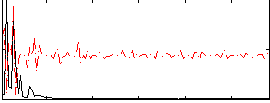
Component:367; Jaccard:0.072984
 |
Component:187; Jaccard:0.072085
 |
Component:053; Jaccard:0.057313
 |
Component:027; Jaccard:0.034398
 |
Component:027; Jaccard:0.013886
 |
Component:048; Jaccard:0.004635
 |
Component:064; Jaccard:0.001719
 |
Correlation:-0.016374
  |
Correlation:0.18215
 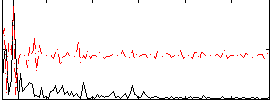 |
Correlation:0.19258
  |
Correlation:0.14119
 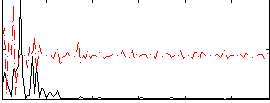 |
Correlation:0.14119
  |
Correlation:0.15513
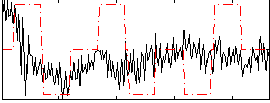 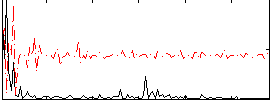 |
Correlation:-0.067766
  |
Cope: 05:"neg_MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
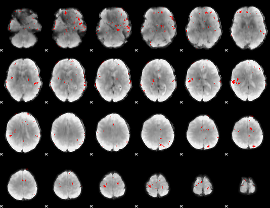 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
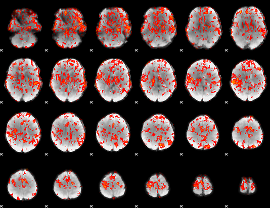 |
GLM Z-score result thresholded by 1.5
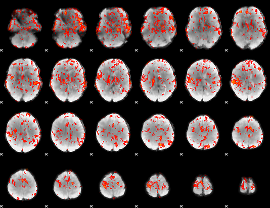 |
GLM Z-score result thresholded by 2
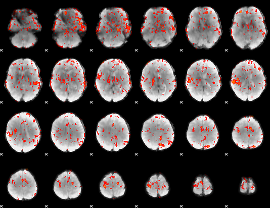 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:084; Jaccard:0.10568
 |
Component:112; Jaccard:0.11427
 |
Component:112; Jaccard:0.12578
 |
Component:112; Jaccard:0.12635
 |
Component:112; Jaccard:0.10725
 |
Component:112; Jaccard:0.074056
 |
Component:266; Jaccard:0.039298
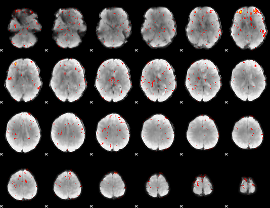 |
Correlation:0.17288
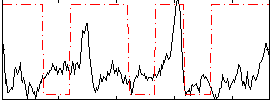 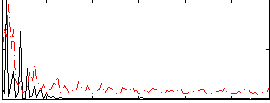 |
Correlation:0.099035
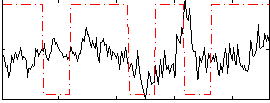  |
Correlation:0.099035
  |
Correlation:0.099035
  |
Correlation:0.099035
  |
Correlation:0.099035
  |
Correlation:0.16294
  |
Component:127; Jaccard:0.099524
 |
Component:127; Jaccard:0.11228
 |
Component:127; Jaccard:0.11765
 |
Component:127; Jaccard:0.10868
 |
Component:256; Jaccard:0.089382
 |
Component:256; Jaccard:0.063953
 |
Component:139; Jaccard:0.036801
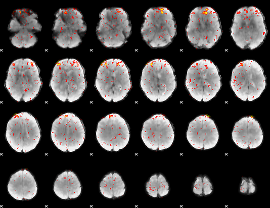 |
Correlation:0.10358
  |
Correlation:0.10358
  |
Correlation:0.10358
  |
Correlation:0.10358
  |
Correlation:0.11421
  |
Correlation:0.11421
  |
Correlation:0.095569
  |
Component:112; Jaccard:0.097476
 |
Component:084; Jaccard:0.1088
 |
Component:084; Jaccard:0.10631
 |
Component:256; Jaccard:0.10163
 |
Component:127; Jaccard:0.087415
 |
Component:139; Jaccard:0.062065
 |
Component:112; Jaccard:0.03644
 |
Correlation:0.099035
  |
Correlation:0.17288
  |
Correlation:0.17288
  |
Correlation:0.11421
  |
Correlation:0.10358
  |
Correlation:0.095569
  |
Correlation:0.099035
  |
Component:354; Jaccard:0.093722
 |
Component:256; Jaccard:0.093392
 |
Component:028; Jaccard:0.10234
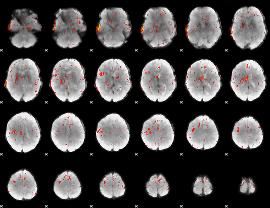 |
Component:028; Jaccard:0.096377
 |
Component:028; Jaccard:0.079947
 |
Component:266; Jaccard:0.0616
 |
Component:256; Jaccard:0.036235
 |
Correlation:-0.049004
 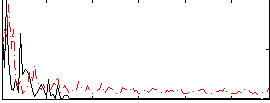 |
Correlation:0.11421
  |
Correlation:8.3216e-05
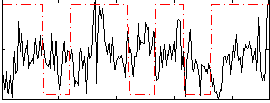 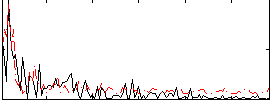 |
Correlation:8.3216e-05
  |
Correlation:8.3216e-05
  |
Correlation:0.16294
  |
Correlation:0.11421
  |
Component:152; Jaccard:0.087123
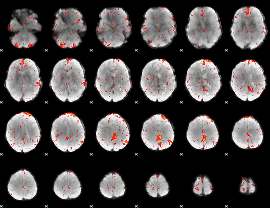 |
Component:028; Jaccard:0.093206
 |
Component:256; Jaccard:0.10209
 |
Component:139; Jaccard:0.092304
 |
Component:139; Jaccard:0.079188
 |
Component:127; Jaccard:0.057731
 |
Component:028; Jaccard:0.030742
 |
Correlation:0.11527
  |
Correlation:8.3216e-05
  |
Correlation:0.11421
  |
Correlation:0.095569
  |
Correlation:0.095569
  |
Correlation:0.10358
  |
Correlation:8.3216e-05
  |
Component:256; Jaccard:0.080218
 |
Component:152; Jaccard:0.091054
 |
Component:139; Jaccard:0.094244
 |
Component:084; Jaccard:0.090272
 |
Component:266; Jaccard:0.077059
 |
Component:028; Jaccard:0.054242
 |
Component:190; Jaccard:0.029952
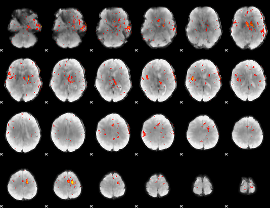 |
Correlation:0.11421
  |
Correlation:0.11527
  |
Correlation:0.095569
  |
Correlation:0.17288
  |
Correlation:0.16294
  |
Correlation:8.3216e-05
  |
Correlation:0.072922
  |
Component:272; Jaccard:0.079594
 |
Component:354; Jaccard:0.089834
 |
Component:097; Jaccard:0.090763
 |
Component:266; Jaccard:0.089022
 |
Component:097; Jaccard:0.072734
 |
Component:190; Jaccard:0.051533
 |
Component:097; Jaccard:0.029577
 |
Correlation:-0.061764
  |
Correlation:-0.049004
  |
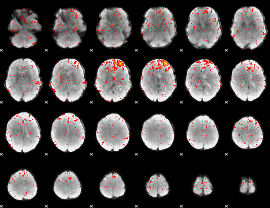
Correlation:0.23997
  |
Correlation:0.16294
  |
Correlation:0.23997
  |
Correlation:0.072922
  |
Correlation:0.23997
  |
Component:028; Jaccard:0.079448
 |
Component:097; Jaccard:0.086448
 |
Component:266; Jaccard:0.088315
 |
Component:097; Jaccard:0.085738
 |
Component:190; Jaccard:0.07214
 |
Component:097; Jaccard:0.051304
 |
Component:127; Jaccard:0.028897
 |
Correlation:8.3216e-05
  |
Correlation:0.23997
  |
Correlation:0.16294
  |
Correlation:0.23997
  |
Correlation:0.072922
  |
Correlation:0.23997
  |
Correlation:0.10358
  |
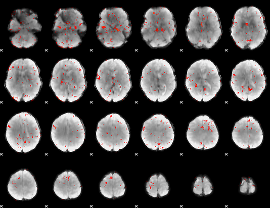
Component:239; Jaccard:0.079329
 |
Component:139; Jaccard:0.086291
 |
Component:272; Jaccard:0.088225
 |
Component:190; Jaccard:0.084455
 |
Component:272; Jaccard:0.067486
 |
Component:104; Jaccard:0.043242
 |
Component:173; Jaccard:0.024402
 |
Correlation:0.036307
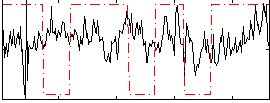 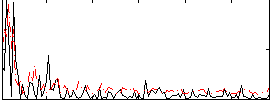 |
Correlation:0.095569
  |
Correlation:-0.061764
  |
Correlation:0.072922
  |
Correlation:-0.061764
  |
Correlation:0.20782
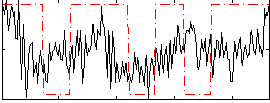 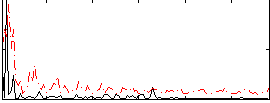 |
Correlation:-0.07049
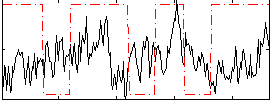 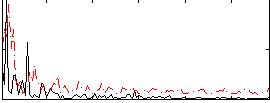 |
Component:097; Jaccard:0.078897
 |
Component:272; Jaccard:0.085805
 |
Component:152; Jaccard:0.08717
 |
Component:272; Jaccard:0.082269
 |
Component:104; Jaccard:0.065
 |
Component:024; Jaccard:0.042337
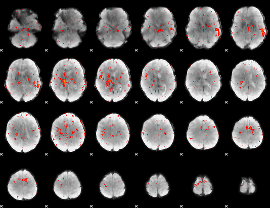 |
Component:104; Jaccard:0.023529
 |
Correlation:0.23997
  |
Correlation:-0.061764
  |
Correlation:0.11527
  |
Correlation:-0.061764
  |
Correlation:0.20782
  |
Correlation:0.16149
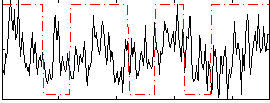  |
Correlation:0.20782
  |
Cope: 06:"neg_REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
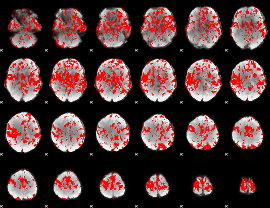 |
GLM binary result thresholded by 1.5
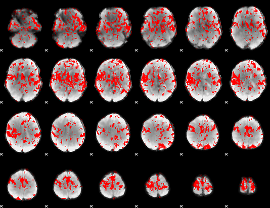 |
GLM binary result thresholded by 2
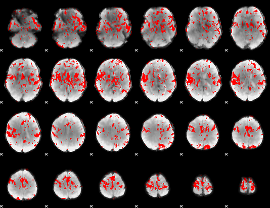 |
GLM binary result thresholded by 2.5
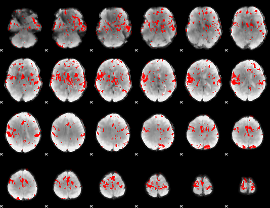 |
GLM binary result thresholded by 3
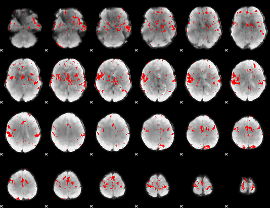 |
GLM binary result thresholded by 3.5
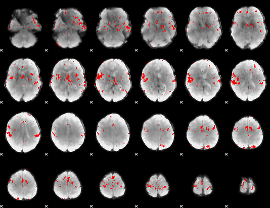 |
GLM binary result thresholded by 4
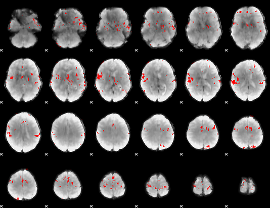 |
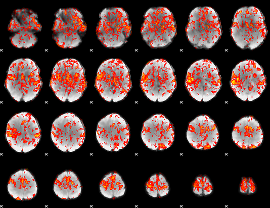
GLM Z-score result thresholded by 1
 |
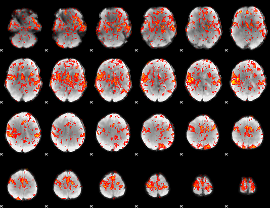
GLM Z-score result thresholded by 1.5
 |
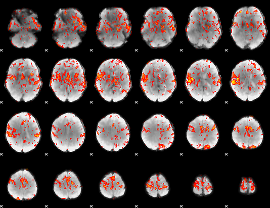
GLM Z-score result thresholded by 2
 |
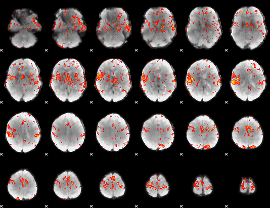
GLM Z-score result thresholded by 2.5
 |
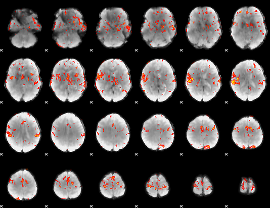
GLM Z-score result thresholded by 3
 |
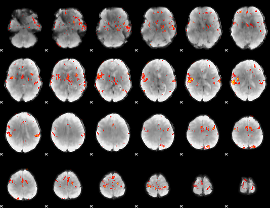
GLM Z-score result thresholded by 3.5
 |
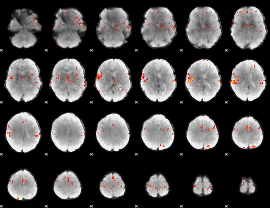
GLM Z-score result thresholded by 4
 |
Component:354; Jaccard:0.12274
 |
Component:354; Jaccard:0.13022
 |
Component:256; Jaccard:0.13578
 |
Component:256; Jaccard:0.15866
 |
Component:256; Jaccard:0.17305
 |
Component:256; Jaccard:0.1705
 |
Component:256; Jaccard:0.15302
 |
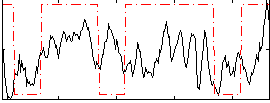
Correlation:0.25142
  |
Correlation:0.25142
  |
Correlation:0.32752
  |
Correlation:0.32752
  |
Correlation:0.32752
  |
Correlation:0.32752
  |
Correlation:0.32752
  |
Component:084; Jaccard:0.11639
 |
Component:084; Jaccard:0.12887
 |
Component:084; Jaccard:0.13521
 |
Component:229; Jaccard:0.14166
 |
Component:229; Jaccard:0.14998
 |
Component:229; Jaccard:0.14286
 |
Component:229; Jaccard:0.11793
 |
Correlation:0.26578
  |
Correlation:0.26578
  |
Correlation:0.26578
  |
Correlation:0.24741
  |
Correlation:0.24741
  |
Correlation:0.24741
  |
Correlation:0.24741
  |
Component:374; Jaccard:0.10381
 |
Component:374; Jaccard:0.11555
 |
Component:354; Jaccard:0.13392
 |
Component:084; Jaccard:0.13419
 |
Component:173; Jaccard:0.1233
 |
Component:190; Jaccard:0.11837
 |
Component:190; Jaccard:0.10388
 |
Correlation:0.10622
  |
Correlation:0.10622
  |
Correlation:0.25142
  |
Correlation:0.26578
  |
Correlation:0.21808
  |
Correlation:0.27795
  |
Correlation:0.27795
  |
Component:256; Jaccard:0.087598
 |
Component:256; Jaccard:0.11017
 |
Component:229; Jaccard:0.12593
 |
Component:354; Jaccard:0.1274
 |
Component:084; Jaccard:0.1201
 |
Component:173; Jaccard:0.1125
 |
Component:173; Jaccard:0.091204
 |
Correlation:0.32752
  |
Correlation:0.32752
  |
Correlation:0.24741
  |
Correlation:0.25142
  |
Correlation:0.26578
  |
Correlation:0.21808
  |
Correlation:0.21808
  |
Component:127; Jaccard:0.087471
 |
Component:229; Jaccard:0.10597
 |
Component:374; Jaccard:0.12316
 |
Component:374; Jaccard:0.12408
 |
Component:190; Jaccard:0.12
 |
Component:374; Jaccard:0.098061
 |
Component:374; Jaccard:0.075466
 |
Correlation:0.19538
  |
Correlation:0.24741
  |
Correlation:0.10622
  |
Correlation:0.10622
  |
Correlation:0.27795
  |
Correlation:0.10622
  |
Correlation:0.10622
  |
Component:173; Jaccard:0.087229
 |
Component:173; Jaccard:0.10162
 |
Component:173; Jaccard:0.1136
 |
Component:173; Jaccard:0.12124
 |
Component:374; Jaccard:0.11463
 |
Component:084; Jaccard:0.095288
 |
Component:084; Jaccard:0.069863
 |
Correlation:0.21808
  |
Correlation:0.21808
  |
Correlation:0.21808
  |
Correlation:0.21808
  |
Correlation:0.10622
  |
Correlation:0.26578
  |
Correlation:0.26578
  |
Component:229; Jaccard:0.086297
 |
Component:127; Jaccard:0.097936
 |
Component:127; Jaccard:0.10397
 |
Component:190; Jaccard:0.11163
 |
Component:354; Jaccard:0.10694
 |
Component:085; Jaccard:0.085016
 |
Component:085; Jaccard:0.067586
 |
Correlation:0.24741
  |
Correlation:0.19538
  |
Correlation:0.19538
  |
Correlation:0.27795
  |
Correlation:0.25142
  |
Correlation:0.10516
  |
Correlation:0.10516
  |
Component:378; Jaccard:0.082268
 |
Component:085; Jaccard:0.08632
 |
Component:190; Jaccard:0.098379
 |
Component:127; Jaccard:0.1038
 |
Component:127; Jaccard:0.094768
 |
Component:354; Jaccard:0.079472
 |
Component:125; Jaccard:0.057664
 |
Correlation:0.15295
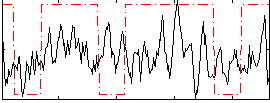 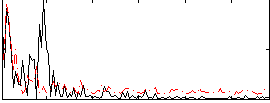 |
Correlation:0.10516
  |
Correlation:0.27795
  |
Correlation:0.19538
  |
Correlation:0.19538
  |
Correlation:0.25142
  |
Correlation:0.23866
  |
Component:085; Jaccard:0.076659
 |
Component:378; Jaccard:0.085198
 |
Component:085; Jaccard:0.093585
 |
Component:085; Jaccard:0.09725
 |
Component:085; Jaccard:0.094604
 |
Component:127; Jaccard:0.078057
 |
Component:127; Jaccard:0.057419
 |
Correlation:0.10516
  |
Correlation:0.15295
  |
Correlation:0.10516
  |
Correlation:0.10516
  |
Correlation:0.10516
  |
Correlation:0.19538
  |
Correlation:0.19538
  |
Component:006; Jaccard:0.075508
 |
Component:190; Jaccard:0.083464
 |
Component:018; Jaccard:0.089208
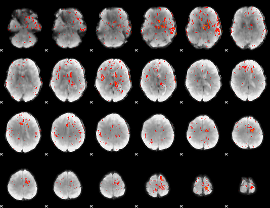 |
Component:018; Jaccard:0.090313
 |
Component:018; Jaccard:0.084348
 |
Component:125; Jaccard:0.073116
 |
Component:039; Jaccard:0.056791
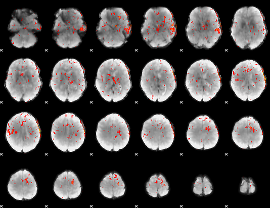 |
Correlation:-0.0022895
  |
Correlation:0.27795
  |
Correlation:0.17071
  |
Correlation:0.17071
  |
Correlation:0.17071
  |
Correlation:0.23866
  |
Correlation:0.17464
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































