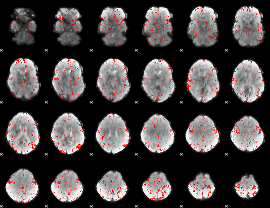
Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"FACES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
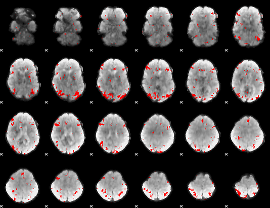 |
GLM binary result thresholded by 3.5
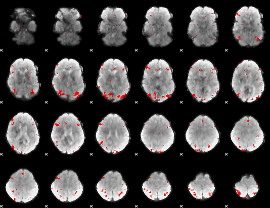 |
GLM binary result thresholded by 4
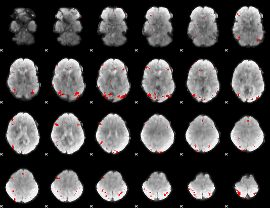 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
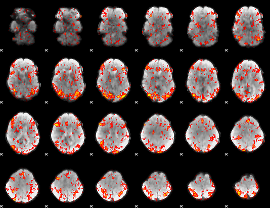 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
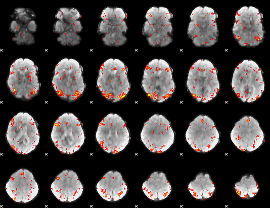 |
GLM Z-score result thresholded by 3
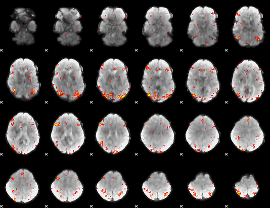 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
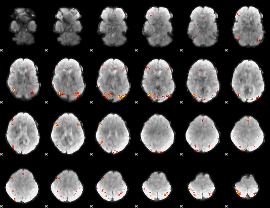 |
Component:021; Jaccard:0.14317
 |
Component:021; Jaccard:0.18868
 |
Component:021; Jaccard:0.23304
 |
Component:021; Jaccard:0.26517
 |
Component:021; Jaccard:0.27123
 |
Component:021; Jaccard:0.24061
 |
Component:021; Jaccard:0.19413
 |
Correlation:0.5493
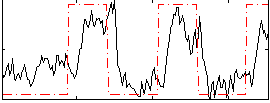  |
Correlation:0.5493
  |
Correlation:0.5493
  |
Correlation:0.5493
  |
Correlation:0.5493
  |
Correlation:0.5493
  |
Correlation:0.5493
  |
Component:156; Jaccard:0.13669
 |
Component:156; Jaccard:0.15699
 |
Component:156; Jaccard:0.16723
 |
Component:353; Jaccard:0.161
 |
Component:270; Jaccard:0.15039
 |
Component:270; Jaccard:0.12452
 |
Component:353; Jaccard:0.10062
 |
Correlation:0.27261
  |
Correlation:0.27261
  |
Correlation:0.27261
  |
Correlation:0.52092
  |
Correlation:0.28481
  |
Correlation:0.28481
  |
Correlation:0.52092
  |
Component:353; Jaccard:0.11914
 |
Component:353; Jaccard:0.14151
 |
Component:353; Jaccard:0.15782
 |
Component:156; Jaccard:0.16058
 |
Component:353; Jaccard:0.14608
 |
Component:353; Jaccard:0.12406
 |
Component:270; Jaccard:0.099491
 |
Correlation:0.52092
  |
Correlation:0.52092
  |
Correlation:0.52092
  |
Correlation:0.27261
  |
Correlation:0.52092
  |
Correlation:0.52092
  |
Correlation:0.28481
  |
Component:270; Jaccard:0.10849
 |
Component:270; Jaccard:0.13412
 |
Component:270; Jaccard:0.15099
 |
Component:270; Jaccard:0.15932
 |
Component:156; Jaccard:0.13611
 |
Component:100; Jaccard:0.10733
 |
Component:100; Jaccard:0.090581
 |
Correlation:0.28481
  |
Correlation:0.28481
  |
Correlation:0.28481
  |
Correlation:0.28481
  |
Correlation:0.27261
  |
Correlation:0.50982
  |
Correlation:0.50982
  |
Component:216; Jaccard:0.10159
 |
Component:216; Jaccard:0.11609
 |
Component:216; Jaccard:0.12333
 |
Component:100; Jaccard:0.12945
 |
Component:100; Jaccard:0.12383
 |
Component:156; Jaccard:0.10534
 |
Component:156; Jaccard:0.078061
 |
Correlation:0.14052
  |
Correlation:0.14052
  |
Correlation:0.14052
  |
Correlation:0.50982
  |
Correlation:0.50982
  |
Correlation:0.27261
  |
Correlation:0.27261
  |
Component:206; Jaccard:0.095386
 |
Component:074; Jaccard:0.10827
 |
Component:100; Jaccard:0.11912
 |
Component:074; Jaccard:0.12037
 |
Component:074; Jaccard:0.11154
 |
Component:074; Jaccard:0.095437
 |
Component:074; Jaccard:0.075669
 |
Correlation:0.08589
  |
Correlation:0.35054
  |
Correlation:0.50982
  |
Correlation:0.35054
  |
Correlation:0.35054
  |
Correlation:0.35054
  |
Correlation:0.35054
  |
Component:074; Jaccard:0.094498
 |
Component:100; Jaccard:0.10384
 |
Component:074; Jaccard:0.11909
 |
Component:216; Jaccard:0.11977
 |
Component:216; Jaccard:0.10459
 |
Component:216; Jaccard:0.084104
 |
Component:216; Jaccard:0.063994
 |
Correlation:0.35054
  |
Correlation:0.50982
  |
Correlation:0.35054
  |
Correlation:0.14052
  |
Correlation:0.14052
  |
Correlation:0.14052
  |
Correlation:0.14052
  |
Component:009; Jaccard:0.091576
 |
Component:206; Jaccard:0.10377
 |
Component:206; Jaccard:0.10664
 |
Component:256; Jaccard:0.10198
 |
Component:256; Jaccard:0.086612
 |
Component:256; Jaccard:0.070436
 |
Component:256; Jaccard:0.056259
 |
Correlation:-0.091883
  |
Correlation:0.08589
  |
Correlation:0.08589
  |
Correlation:0.1844
  |
Correlation:0.1844
  |
Correlation:0.1844
  |
Correlation:0.1844
  |
Component:091; Jaccard:0.089443
 |
Component:256; Jaccard:0.098111
 |
Component:256; Jaccard:0.10369
 |
Component:206; Jaccard:0.098464
 |
Component:206; Jaccard:0.083836
 |
Component:206; Jaccard:0.065824
 |
Component:206; Jaccard:0.048758
 |
Correlation:0.2266
  |
Correlation:0.1844
  |
Correlation:0.1844
  |
Correlation:0.08589
  |
Correlation:0.08589
  |
Correlation:0.08589
  |
Correlation:0.08589
  |
Component:256; Jaccard:0.087663
 |
Component:388; Jaccard:0.09467
 |
Component:388; Jaccard:0.097547
 |
Component:388; Jaccard:0.094304
 |
Component:388; Jaccard:0.082202
 |
Component:388; Jaccard:0.064164
 |
Component:388; Jaccard:0.047463
 |
Correlation:0.1844
  |
Correlation:0.34599
  |
Correlation:0.34599
  |
Correlation:0.34599
  |
Correlation:0.34599
  |
Correlation:0.34599
  |
Correlation:0.34599
  |
Cope: 02:"SHAPES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
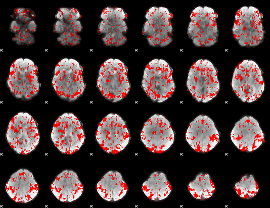 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
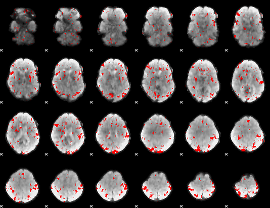 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
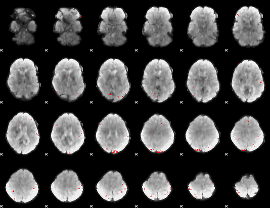 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
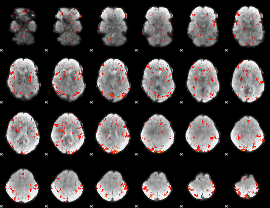 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
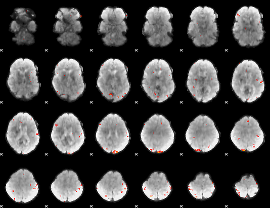 |
GLM Z-score result thresholded by 4
 |
Component:009; Jaccard:0.1421
 |
Component:216; Jaccard:0.16809
 |
Component:216; Jaccard:0.18371
 |
Component:216; Jaccard:0.16407
 |
Component:216; Jaccard:0.12201
 |
Component:216; Jaccard:0.078375
 |
Component:216; Jaccard:0.04729
 |
Correlation:0.24509
  |
Correlation:0.037515
  |
Correlation:0.037515
  |
Correlation:0.037515
  |
Correlation:0.037515
  |
Correlation:0.037515
  |
Correlation:0.037515
  |
Component:216; Jaccard:0.13749
 |
Component:009; Jaccard:0.15568
 |
Component:009; Jaccard:0.14947
 |
Component:270; Jaccard:0.13001
 |
Component:270; Jaccard:0.10134
 |
Component:270; Jaccard:0.063286
 |
Component:270; Jaccard:0.037443
 |
Correlation:0.037515
  |
Correlation:0.24509
  |
Correlation:0.24509
  |
Correlation:-0.029717
  |
Correlation:-0.029717
  |
Correlation:-0.029717
  |
Correlation:-0.029717
  |
Component:270; Jaccard:0.11354
 |
Component:270; Jaccard:0.13625
 |
Component:270; Jaccard:0.14665
 |
Component:009; Jaccard:0.11713
 |
Component:009; Jaccard:0.077053
 |
Component:009; Jaccard:0.046598
 |
Component:009; Jaccard:0.024364
 |
Correlation:-0.029717
  |
Correlation:-0.029717
  |
Correlation:-0.029717
  |
Correlation:0.24509
  |
Correlation:0.24509
  |
Correlation:0.24509
  |
Correlation:0.24509
  |
Component:206; Jaccard:0.092269
 |
Component:206; Jaccard:0.096001
 |
Component:206; Jaccard:0.089835
 |
Component:206; Jaccard:0.071408
 |
Component:358; Jaccard:0.051242
 |
Component:358; Jaccard:0.031884
 |
Component:360; Jaccard:0.019572
 |
Correlation:-0.1447
  |
Correlation:-0.1447
  |
Correlation:-0.1447
  |
Correlation:-0.1447
  |
Correlation:-0.043921
  |
Correlation:-0.043921
  |
Correlation:0.39558
  |
Component:156; Jaccard:0.09171
 |
Component:156; Jaccard:0.091259
 |
Component:358; Jaccard:0.082861
 |
Component:358; Jaccard:0.070726
 |
Component:206; Jaccard:0.047159
 |
Component:074; Jaccard:0.030854
 |
Component:358; Jaccard:0.018065
 |
Correlation:-0.25628
  |
Correlation:-0.25628
  |
Correlation:-0.043921
  |
Correlation:-0.043921
  |
Correlation:-0.1447
  |
Correlation:-0.24808
  |
Correlation:-0.043921
  |
Component:358; Jaccard:0.081107
 |
Component:358; Jaccard:0.08523
 |
Component:256; Jaccard:0.080652
 |
Component:256; Jaccard:0.06484
 |
Component:353; Jaccard:0.046352
 |
Component:353; Jaccard:0.028936
 |
Component:256; Jaccard:0.017968
 |
Correlation:-0.043921
  |
Correlation:-0.043921
  |
Correlation:-0.022993
  |
Correlation:-0.022993
  |
Correlation:-0.34005
  |
Correlation:-0.34005
  |
Correlation:-0.022993
  |
Component:353; Jaccard:0.080821
 |
Component:256; Jaccard:0.084188
 |
Component:156; Jaccard:0.077558
 |
Component:353; Jaccard:0.063775
 |
Component:074; Jaccard:0.044782
 |
Component:256; Jaccard:0.028286
 |
Component:353; Jaccard:0.01696
 |
Correlation:-0.34005
  |
Correlation:-0.022993
  |
Correlation:-0.25628
  |
Correlation:-0.34005
  |
Correlation:-0.24808
  |
Correlation:-0.022993
  |
Correlation:-0.34005
  |
Component:256; Jaccard:0.079817
 |
Component:353; Jaccard:0.082794
 |
Component:353; Jaccard:0.077298
 |
Component:074; Jaccard:0.061572
 |
Component:256; Jaccard:0.043504
 |
Component:206; Jaccard:0.028114
 |
Component:074; Jaccard:0.016935
 |
Correlation:-0.022993
  |
Correlation:-0.34005
  |
Correlation:-0.34005
  |
Correlation:-0.24808
  |
Correlation:-0.022993
  |
Correlation:-0.1447
  |
Correlation:-0.24808
  |
Component:126; Jaccard:0.073934
 |
Component:074; Jaccard:0.074007
 |
Component:074; Jaccard:0.070975
 |
Component:156; Jaccard:0.056249
 |
Component:039; Jaccard:0.036991
 |
Component:360; Jaccard:0.027478
 |
Component:206; Jaccard:0.015859
 |
Correlation:-0.12489
  |
Correlation:-0.24808
  |
Correlation:-0.24808
  |
Correlation:-0.25628
  |
Correlation:0.19615
  |
Correlation:0.39558
  |
Correlation:-0.1447
  |
Component:078; Jaccard:0.072693
 |
Component:078; Jaccard:0.070779
 |
Component:039; Jaccard:0.065456
 |
Component:039; Jaccard:0.054341
 |
Component:156; Jaccard:0.035776
 |
Component:021; Jaccard:0.020909
 |
Component:021; Jaccard:0.011743
 |
Correlation:-0.32413
  |
Correlation:-0.32413
  |
Correlation:0.19615
  |
Correlation:0.19615
  |
Correlation:-0.25628
  |
Correlation:-0.52494
  |
Correlation:-0.52494
  |
Cope: 03:"FACES-SHAPES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
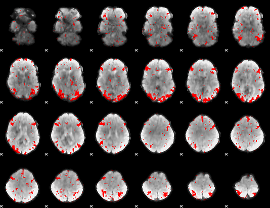 |
GLM binary result thresholded by 3
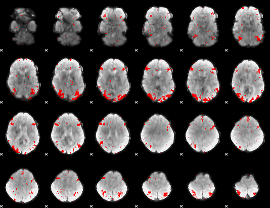 |
GLM binary result thresholded by 3.5
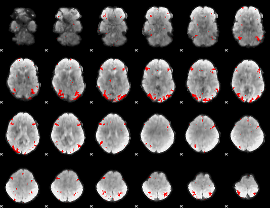 |
GLM binary result thresholded by 4
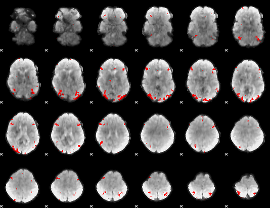 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
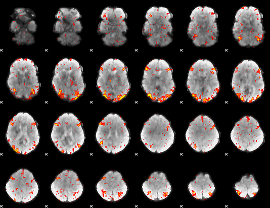 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:021; Jaccard:0.14046
 |
Component:021; Jaccard:0.20038
 |
Component:021; Jaccard:0.27445
 |
Component:021; Jaccard:0.32885
 |
Component:021; Jaccard:0.35045
 |
Component:021; Jaccard:0.31993
 |
Component:021; Jaccard:0.26774
 |
Correlation:0.58268
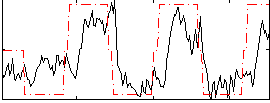  |
Correlation:0.58268
  |
Correlation:0.58268
  |
Correlation:0.58268
  |
Correlation:0.58268
  |
Correlation:0.58268
  |
Correlation:0.58268
  |
Component:156; Jaccard:0.11901
 |
Component:100; Jaccard:0.1663
 |
Component:100; Jaccard:0.22459
 |
Component:100; Jaccard:0.27129
 |
Component:100; Jaccard:0.2794
 |
Component:100; Jaccard:0.26102
 |
Component:100; Jaccard:0.21569
 |
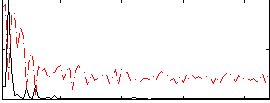
Correlation:0.28688
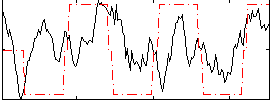  |
Correlation:0.46041
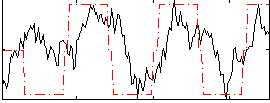 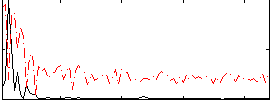 |
Correlation:0.46041
  |
Correlation:0.46041
  |
Correlation:0.46041
  |
Correlation:0.46041
  |
Correlation:0.46041
  |
Component:100; Jaccard:0.11774
 |
Component:156; Jaccard:0.14249
 |
Component:388; Jaccard:0.16117
 |
Component:388; Jaccard:0.16209
 |
Component:388; Jaccard:0.14416
 |
Component:353; Jaccard:0.12321
 |
Component:353; Jaccard:0.09794
 |
Correlation:0.46041
  |
Correlation:0.28688
  |
Correlation:0.34511
  |
Correlation:0.34511
  |
Correlation:0.34511
  |
Correlation:0.467
  |
Correlation:0.467
  |
Component:388; Jaccard:0.11745
 |
Component:388; Jaccard:0.14207
 |
Component:156; Jaccard:0.15656
 |
Component:156; Jaccard:0.1557
 |
Component:353; Jaccard:0.14259
 |
Component:388; Jaccard:0.11758
 |
Component:388; Jaccard:0.089768
 |
Correlation:0.34511
  |
Correlation:0.34511
  |
Correlation:0.28688
  |
Correlation:0.28688
  |
Correlation:0.467
  |
Correlation:0.34511
  |
Correlation:0.34511
  |
Component:353; Jaccard:0.10595
 |
Component:353; Jaccard:0.12687
 |
Component:353; Jaccard:0.1444
 |
Component:353; Jaccard:0.14926
 |
Component:156; Jaccard:0.13825
 |
Component:156; Jaccard:0.11156
 |
Component:156; Jaccard:0.084731
 |
Correlation:0.467
  |
Correlation:0.467
  |
Correlation:0.467
  |
Correlation:0.467
  |
Correlation:0.28688
  |
Correlation:0.28688
  |
Correlation:0.28688
  |
Component:008; Jaccard:0.080894
 |
Component:074; Jaccard:0.091614
 |
Component:074; Jaccard:0.099461
 |
Component:288; Jaccard:0.10069
 |
Component:288; Jaccard:0.092688
 |
Component:288; Jaccard:0.083531
 |
Component:288; Jaccard:0.068717
 |
Correlation:0.43889
  |
Correlation:0.3247
  |
Correlation:0.3247
  |
Correlation:0.42699
  |
Correlation:0.42699
  |
Correlation:0.42699
  |
Correlation:0.42699
  |
Component:074; Jaccard:0.079133
 |
Component:288; Jaccard:0.09151
 |
Component:288; Jaccard:0.099377
 |
Component:074; Jaccard:0.097382
 |
Component:074; Jaccard:0.090098
 |
Component:074; Jaccard:0.078023
 |
Component:074; Jaccard:0.063689
 |
Correlation:0.3247
  |
Correlation:0.42699
  |
Correlation:0.42699
  |
Correlation:0.3247
  |
Correlation:0.3247
  |
Correlation:0.3247
  |
Correlation:0.3247
  |
Component:023; Jaccard:0.078786
 |
Component:008; Jaccard:0.084449
 |
Component:008; Jaccard:0.08197
 |
Component:270; Jaccard:0.075067
 |
Component:264; Jaccard:0.068731
 |
Component:264; Jaccard:0.061299
 |
Component:264; Jaccard:0.054243
 |
Correlation:0.34127
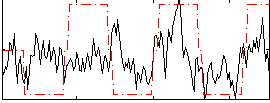  |
Correlation:0.43889
  |
Correlation:0.43889
  |
Correlation:0.1706
  |
Correlation:0.18122
 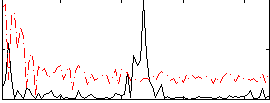 |
Correlation:0.18122
  |
Correlation:0.18122
  |
Component:288; Jaccard:0.078388
 |
Component:023; Jaccard:0.08182
 |
Component:091; Jaccard:0.078224
 |
Component:008; Jaccard:0.073176
 |
Component:270; Jaccard:0.06777
 |
Component:270; Jaccard:0.056886
 |
Component:270; Jaccard:0.04444
 |
Correlation:0.42699
  |
Correlation:0.34127
  |
Correlation:0.2257
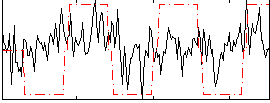  |
Correlation:0.43889
  |
Correlation:0.1706
  |
Correlation:0.1706
  |
Correlation:0.1706
  |
Component:395; Jaccard:0.07818
 |
Component:091; Jaccard:0.080735
 |
Component:023; Jaccard:0.077926
 |
Component:264; Jaccard:0.072585
 |
Component:370; Jaccard:0.06414
 |
Component:256; Jaccard:0.053809
 |
Component:256; Jaccard:0.042766
 |
Correlation:0.19907
  |
Correlation:0.2257
  |
Correlation:0.34127
  |
Correlation:0.18122
  |
Correlation:0.34454
  |
Correlation:0.11249
  |
Correlation:0.11249
  |
Cope: 04:"neg_FACES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
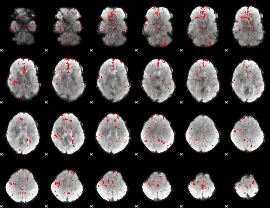 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
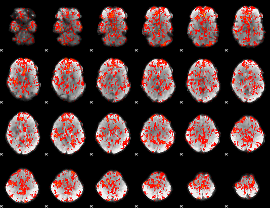 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
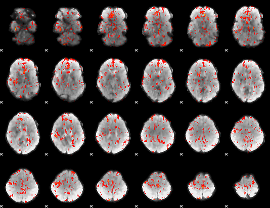 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
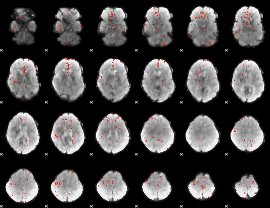 |
GLM Z-score result thresholded by 3.5
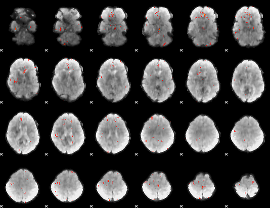 |
GLM Z-score result thresholded by 4
 |
Component:106; Jaccard:0.11164
 |
Component:106; Jaccard:0.12419
 |
Component:106; Jaccard:0.12635
 |
Component:106; Jaccard:0.10281
 |
Component:106; Jaccard:0.067258
 |
Component:106; Jaccard:0.036125
 |
Component:266; Jaccard:0.018591
 |
Correlation:0.2438
  |
Correlation:0.2438
  |
Correlation:0.2438
  |
Correlation:0.2438
  |
Correlation:0.2438
  |
Correlation:0.2438
  |
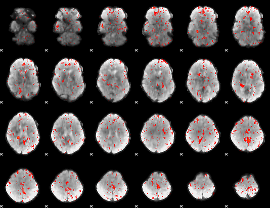
Correlation:0.13339
  |
Component:181; Jaccard:0.10691
 |
Component:181; Jaccard:0.11292
 |
Component:181; Jaccard:0.10273
 |
Component:029; Jaccard:0.086292
 |
Component:266; Jaccard:0.06211
 |
Component:266; Jaccard:0.035179
 |
Component:355; Jaccard:0.016817
 |
Correlation:-0.094588
  |
Correlation:-0.094588
  |
Correlation:-0.094588
  |
Correlation:-0.058723
  |
Correlation:0.13339
  |
Correlation:0.13339
  |
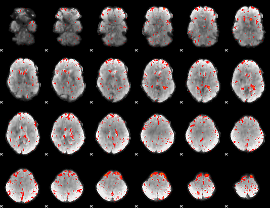
Correlation:0.44737
  |
Component:029; Jaccard:0.090405
 |
Component:029; Jaccard:0.10096
 |
Component:029; Jaccard:0.10233
 |
Component:266; Jaccard:0.083412
 |
Component:029; Jaccard:0.060928
 |
Component:029; Jaccard:0.032963
 |
Component:106; Jaccard:0.01565
 |
Correlation:-0.058723
  |
Correlation:-0.058723
  |
Correlation:-0.058723
  |
Correlation:0.13339
  |
Correlation:-0.058723
  |
Correlation:-0.058723
  |
Correlation:0.2438
  |
Component:059; Jaccard:0.088915
 |
Component:059; Jaccard:0.095093
 |
Component:266; Jaccard:0.090127
 |
Component:181; Jaccard:0.078738
 |
Component:153; Jaccard:0.044217
 |
Component:080; Jaccard:0.026468
 |
Component:080; Jaccard:0.01546
 |
Correlation:-0.080624
  |
Correlation:-0.080624
  |
Correlation:0.13339
  |
Correlation:-0.094588
  |
Correlation:-0.089981
  |
Correlation:0.31848
  |
Correlation:0.31848
  |
Component:153; Jaccard:0.08326
 |
Component:153; Jaccard:0.087479
 |
Component:059; Jaccard:0.088769
 |
Component:059; Jaccard:0.067063
 |
Component:181; Jaccard:0.043054
 |
Component:355; Jaccard:0.02545
 |
Component:029; Jaccard:0.015291
 |
Correlation:-0.089981
  |
Correlation:-0.089981
  |
Correlation:-0.080624
  |
Correlation:-0.080624
  |
Correlation:-0.094588
  |
Correlation:0.44737
  |
Correlation:-0.058723
  |
Component:283; Jaccard:0.077861
 |
Component:266; Jaccard:0.086952
 |
Component:153; Jaccard:0.08159
 |
Component:153; Jaccard:0.066111
 |
Component:080; Jaccard:0.04228
 |
Component:153; Jaccard:0.024114
 |
Component:265; Jaccard:0.011492
 |
Correlation:-0.010174
  |
Correlation:0.13339
  |
Correlation:-0.089981
  |
Correlation:-0.089981
  |
Correlation:0.31848
  |
Correlation:-0.089981
  |
Correlation:0.042192
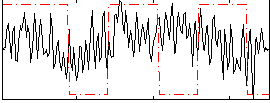  |
Component:266; Jaccard:0.077304
 |
Component:244; Jaccard:0.075629
 |
Component:244; Jaccard:0.074768
 |
Component:244; Jaccard:0.063718
 |
Component:244; Jaccard:0.041045
 |
Component:228; Jaccard:0.022787
 |
Component:228; Jaccard:0.011164
 |
Correlation:0.13339
  |
Correlation:-0.039882
  |
Correlation:-0.039882
  |
Correlation:-0.039882
  |
Correlation:-0.039882
  |
Correlation:0.20407
 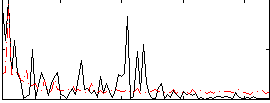 |
Correlation:0.20407
  |
Component:254; Jaccard:0.07501
 |
Component:254; Jaccard:0.075277
 |
Component:327; Jaccard:0.070231
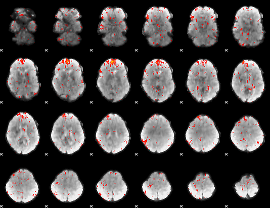 |
Component:327; Jaccard:0.057176
 |
Component:059; Jaccard:0.039021
 |
Component:251; Jaccard:0.022443
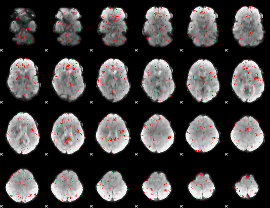 |
Component:153; Jaccard:0.010666
 |
Correlation:0.0007268
 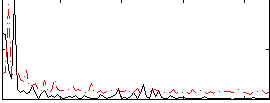 |
Correlation:0.0007268
  |
Correlation:0.060414
 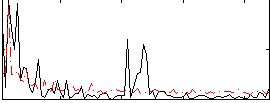 |
Correlation:0.060414
  |
Correlation:-0.080624
  |
Correlation:0.12684
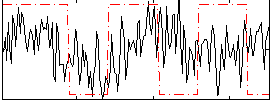 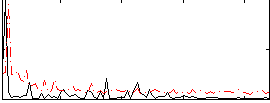 |
Correlation:-0.089981
  |
Component:293; Jaccard:0.073019
 |
Component:293; Jaccard:0.07489
 |
Component:254; Jaccard:0.068755
 |
Component:228; Jaccard:0.055229
 |
Component:228; Jaccard:0.038635
 |
Component:034; Jaccard:0.02196
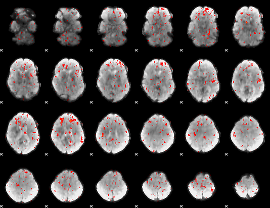 |
Component:251; Jaccard:0.01015
 |
Correlation:0.056492
 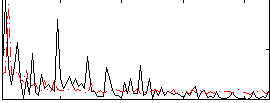 |
Correlation:0.056492
  |
Correlation:0.0007268
  |
Correlation:0.20407
  |
Correlation:0.20407
  |
Correlation:0.12615
  |
Correlation:0.12684
  |
Component:244; Jaccard:0.072282
 |
Component:283; Jaccard:0.073808
 |
Component:293; Jaccard:0.068645
 |
Component:254; Jaccard:0.05515
 |
Component:355; Jaccard:0.037572
 |
Component:341; Jaccard:0.021355
 |
Component:034; Jaccard:0.00974
 |
Correlation:-0.039882
  |
Correlation:-0.010174
  |
Correlation:0.056492
  |
Correlation:0.0007268
  |
Correlation:0.44737
  |
Correlation:0.37958
 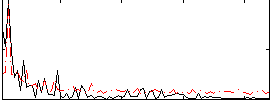 |
Correlation:0.12615
  |
Cope: 05:"neg_SHAPES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
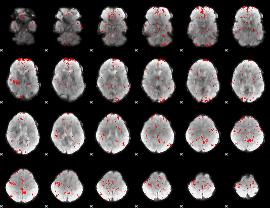 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
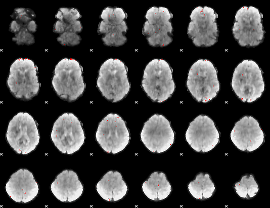 |
GLM Z-score result thresholded by 1
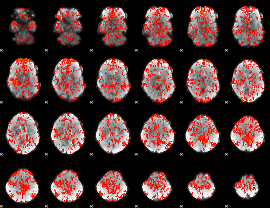 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
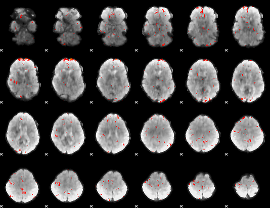 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:181; Jaccard:0.12891
 |
Component:181; Jaccard:0.15056
 |
Component:181; Jaccard:0.15807
 |
Component:181; Jaccard:0.13505
 |
Component:181; Jaccard:0.08868
 |
Component:181; Jaccard:0.042983
 |
Component:181; Jaccard:0.018656
 |
Correlation:0.33897
  |
Correlation:0.33897
  |
Correlation:0.33897
  |
Correlation:0.33897
  |
Correlation:0.33897
  |
Correlation:0.33897
  |
Correlation:0.33897
  |
Component:090; Jaccard:0.1053
 |
Component:059; Jaccard:0.12003
 |
Component:059; Jaccard:0.12528
 |
Component:059; Jaccard:0.10739
 |
Component:059; Jaccard:0.068415
 |
Component:059; Jaccard:0.032453
 |
Component:055; Jaccard:0.013947
 |
Correlation:0.37568
  |
Correlation:0.37377
  |
Correlation:0.37377
  |
Correlation:0.37377
  |
Correlation:0.37377
  |
Correlation:0.37377
  |
Correlation:0.26827
  |
Component:059; Jaccard:0.10291
 |
Component:090; Jaccard:0.1132
 |
Component:090; Jaccard:0.10882
 |
Component:090; Jaccard:0.086557
 |
Component:090; Jaccard:0.056696
 |
Component:090; Jaccard:0.029789
 |
Component:090; Jaccard:0.013825
 |
Correlation:0.37377
  |
Correlation:0.37568
  |
Correlation:0.37568
  |
Correlation:0.37568
  |
Correlation:0.37568
  |
Correlation:0.37568
  |
Correlation:0.37568
  |
Component:106; Jaccard:0.090641
 |
Component:106; Jaccard:0.099771
 |
Component:106; Jaccard:0.10048
 |
Component:106; Jaccard:0.082906
 |
Component:106; Jaccard:0.05146
 |
Component:055; Jaccard:0.028148
 |
Component:250; Jaccard:0.013769
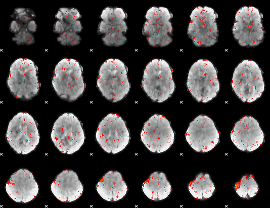 |
Correlation:0.10819
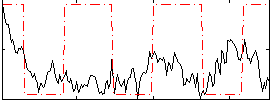  |
Correlation:0.10819
  |
Correlation:0.10819
  |
Correlation:0.10819
  |
Correlation:0.10819
  |
Correlation:0.26827
  |
Correlation:0.34318
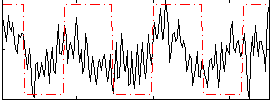 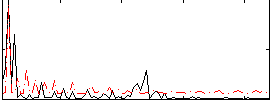 |
Component:371; Jaccard:0.08905
 |
Component:371; Jaccard:0.094174
 |
Component:055; Jaccard:0.08837
 |
Component:055; Jaccard:0.075442
 |
Component:250; Jaccard:0.049637
 |
Component:250; Jaccard:0.0279
 |
Component:059; Jaccard:0.013679
 |
Correlation:0.19787
  |
Correlation:0.19787
  |
Correlation:0.26827
  |
Correlation:0.26827
  |
Correlation:0.34318
  |
Correlation:0.34318
  |
Correlation:0.37377
  |
Component:391; Jaccard:0.082446
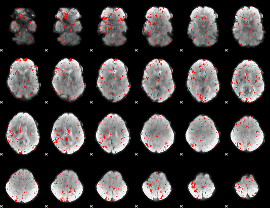 |
Component:391; Jaccard:0.088333
 |
Component:371; Jaccard:0.08726
 |
Component:391; Jaccard:0.071366
 |
Component:055; Jaccard:0.049021
 |
Component:391; Jaccard:0.02568
 |
Component:391; Jaccard:0.011992
 |
Correlation:0.39567
  |
Correlation:0.39567
  |
Correlation:0.19787
  |
Correlation:0.39567
  |
Correlation:0.26827
  |
Correlation:0.39567
  |
Correlation:0.39567
  |
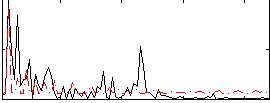
Component:389; Jaccard:0.081854
 |
Component:055; Jaccard:0.087197
 |
Component:391; Jaccard:0.085104
 |
Component:250; Jaccard:0.068885
 |
Component:391; Jaccard:0.047003
 |
Component:106; Jaccard:0.023585
 |
Component:106; Jaccard:0.010558
 |
Correlation:0.56888
  |
Correlation:0.26827
  |
Correlation:0.39567
  |
Correlation:0.34318
  |
Correlation:0.39567
  |
Correlation:0.10819
  |
Correlation:0.10819
  |
Component:263; Jaccard:0.080862
 |
Component:389; Jaccard:0.085806
 |
Component:389; Jaccard:0.080137
 |
Component:371; Jaccard:0.066539
 |
Component:371; Jaccard:0.039263
 |
Component:371; Jaccard:0.019066
 |
Component:100; Jaccard:0.008223
 |
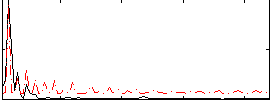
Correlation:0.15967
  |
Correlation:0.56888
  |
Correlation:0.56888
  |
Correlation:0.19787
  |
Correlation:0.19787
  |
Correlation:0.19787
  |
Correlation:0.33899
  |
Component:055; Jaccard:0.080476
 |
Component:263; Jaccard:0.084121
 |
Component:029; Jaccard:0.079646
 |
Component:029; Jaccard:0.062621
 |
Component:029; Jaccard:0.039128
 |
Component:389; Jaccard:0.018739
 |
Component:069; Jaccard:0.007891
 |
Correlation:0.26827
  |
Correlation:0.15967
  |
Correlation:0.028831
  |
Correlation:0.028831
  |
Correlation:0.028831
  |
Correlation:0.56888
  |
Correlation:0.15566
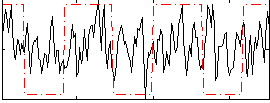 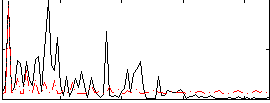 |
Component:029; Jaccard:0.075268
 |
Component:029; Jaccard:0.082305
 |
Component:263; Jaccard:0.079448
 |
Component:389; Jaccard:0.06122
 |
Component:389; Jaccard:0.037902
 |
Component:029; Jaccard:0.018501
 |
Component:080; Jaccard:0.007441
 |
Correlation:0.028831
  |
Correlation:0.028831
  |
Correlation:0.15967
  |
Correlation:0.56888
  |
Correlation:0.56888
  |
Correlation:0.028831
  |
Correlation:-0.14381
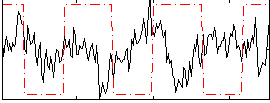 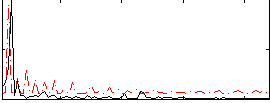 |
Cope: 06:"SHAPES-FACES" BACK
|
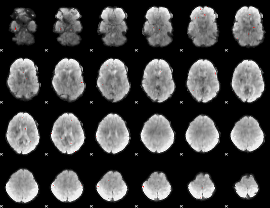
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
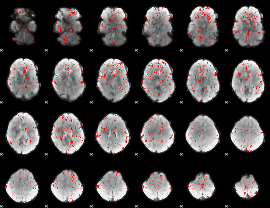 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
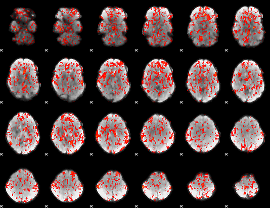
GLM binary result thresholded by 4
 |
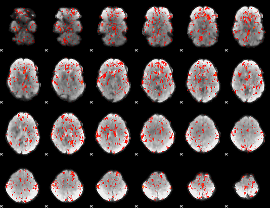
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:155; Jaccard:0.087012
 |
Component:341; Jaccard:0.084865
 |
Component:341; Jaccard:0.082397
 |
Component:341; Jaccard:0.063065
 |
Component:341; Jaccard:0.038651
 |
Component:341; Jaccard:0.018592
 |
Component:355; Jaccard:0.008238
 |
Correlation:0.14768
  |
Correlation:0.32301
 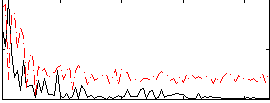 |
Correlation:0.32301
  |
Correlation:0.32301
  |
Correlation:0.32301
  |
Correlation:0.32301
  |
Correlation:0.45993
 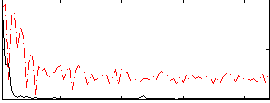 |
Component:106; Jaccard:0.077325
 |
Component:155; Jaccard:0.08369
 |
Component:093; Jaccard:0.07319
 |
Component:093; Jaccard:0.051093
 |
Component:355; Jaccard:0.030544
 |
Component:355; Jaccard:0.018217
 |
Component:341; Jaccard:0.007194
 |
Correlation:0.073556
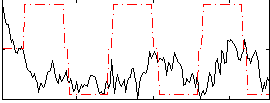  |
Correlation:0.14768
  |
Correlation:0.16958
  |
Correlation:0.16958
  |
Correlation:0.45993
  |
Correlation:0.45993
  |
Correlation:0.32301
  |
Component:341; Jaccard:0.075714
 |
Component:093; Jaccard:0.078328
 |
Component:155; Jaccard:0.067858
 |
Component:234; Jaccard:0.044324
 |
Component:093; Jaccard:0.026125
 |
Component:234; Jaccard:0.01209
 |
Component:266; Jaccard:0.005312
 |
Correlation:0.32301
  |
Correlation:0.16958
  |
Correlation:0.14768
  |
Correlation:0.32571
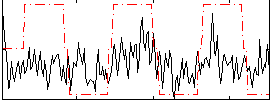 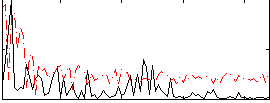 |
Correlation:0.16958
  |
Correlation:0.32571
  |
Correlation:0.12983
  |
Component:093; Jaccard:0.074371
 |
Component:106; Jaccard:0.074205
 |
Component:234; Jaccard:0.063282
 |
Component:355; Jaccard:0.043061
 |
Component:234; Jaccard:0.025552
 |
Component:266; Jaccard:0.011954
 |
Component:130; Jaccard:0.005276
 |
Correlation:0.16958
  |
Correlation:0.073556
  |
Correlation:0.32571
  |
Correlation:0.45993
  |
Correlation:0.32571
  |
Correlation:0.12983
  |
Correlation:0.27048
  |
Component:123; Jaccard:0.073344
 |
Component:234; Jaccard:0.068983
 |
Component:266; Jaccard:0.057991
 |
Component:155; Jaccard:0.042298
 |
Component:266; Jaccard:0.022351
 |
Component:396; Jaccard:0.010651
 |
Component:396; Jaccard:0.004385
 |
Correlation:0.067858
  |
Correlation:0.32571
  |
Correlation:0.12983
  |
Correlation:0.14768
  |
Correlation:0.12983
  |
Correlation:0.22725
  |
Correlation:0.22725
  |
Component:317; Jaccard:0.072662
 |
Component:029; Jaccard:0.068081
 |
Component:106; Jaccard:0.056395
 |
Component:266; Jaccard:0.041804
 |
Component:396; Jaccard:0.020178
 |
Component:093; Jaccard:0.01052
 |
Component:093; Jaccard:0.00415
 |
Correlation:-0.0090426
  |
Correlation:-0.04749
  |
Correlation:0.073556
  |
Correlation:0.12983
  |
Correlation:0.22725
  |
Correlation:0.16958
  |
Correlation:0.16958
  |
Component:029; Jaccard:0.070893
 |
Component:266; Jaccard:0.066519
 |
Component:029; Jaccard:0.056002
 |
Component:169; Jaccard:0.036611
 |
Component:155; Jaccard:0.019237
 |
Component:130; Jaccard:0.009698
 |
Component:317; Jaccard:0.003598
 |
Correlation:-0.04749
  |
Correlation:0.12983
  |
Correlation:-0.04749
  |
Correlation:0.36616
  |
Correlation:0.14768
  |
Correlation:0.27048
  |
Correlation:-0.0090426
  |
Component:234; Jaccard:0.068148
 |
Component:123; Jaccard:0.065701
 |
Component:355; Jaccard:0.051988
 |
Component:029; Jaccard:0.036566
 |
Component:367; Jaccard:0.018986
 |
Component:367; Jaccard:0.009132
 |
Component:169; Jaccard:0.003567
 |
Correlation:0.32571
  |
Correlation:0.067858
  |
Correlation:0.45993
  |
Correlation:-0.04749
  |
Correlation:0.089648
  |
Correlation:0.089648
  |
Correlation:0.36616
  |
Component:295; Jaccard:0.068073
 |
Component:317; Jaccard:0.064585
 |
Component:169; Jaccard:0.051785
 |
Component:317; Jaccard:0.036062
 |
Component:169; Jaccard:0.018853
 |
Component:155; Jaccard:0.008898
 |
Component:316; Jaccard:0.003566
 |
Correlation:-0.13419
  |
Correlation:-0.0090426
  |
Correlation:0.36616
  |
Correlation:-0.0090426
  |
Correlation:0.36616
  |
Correlation:0.14768
  |
Correlation:0.038598
  |
Component:346; Jaccard:0.067216
 |
Component:302; Jaccard:0.062381
 |
Component:317; Jaccard:0.051604
 |
Component:396; Jaccard:0.035797
 |
Component:346; Jaccard:0.018078
 |
Component:169; Jaccard:0.008557
 |
Component:367; Jaccard:0.003528
 |
Correlation:-0.042652
  |
Correlation:-0.019177
  |
Correlation:-0.0090426
  |
Correlation:0.22725
  |
Correlation:-0.042652
  |
Correlation:0.36616
  |
Correlation:0.089648
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































