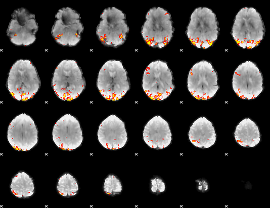
Task name: EMOTION, TR=0.72, totally 176 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"FACES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
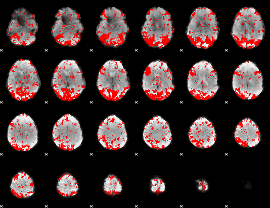 |
GLM binary result thresholded by 2
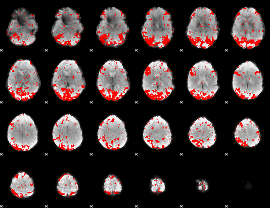 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
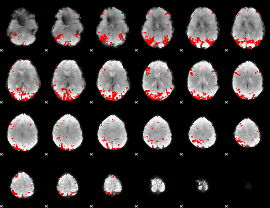 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
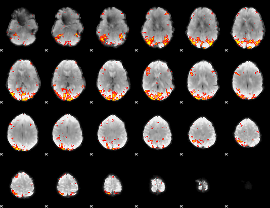 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:205; Jaccard:0.12907
 |
Component:089; Jaccard:0.1582
 |
Component:089; Jaccard:0.19836
 |
Component:089; Jaccard:0.23275
 |
Component:089; Jaccard:0.25532
 |
Component:089; Jaccard:0.25564
 |
Component:353; Jaccard:0.24692
 |
Correlation:0.34523
  |
Correlation:0.57654
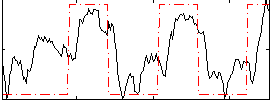 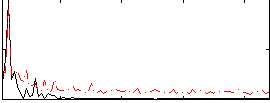 |
Correlation:0.57654
  |
Correlation:0.57654
  |
Correlation:0.57654
  |
Correlation:0.57654
  |
Correlation:0.26442
  |
Component:069; Jaccard:0.12184
 |
Component:205; Jaccard:0.15714
 |
Component:353; Jaccard:0.19386
 |
Component:353; Jaccard:0.22588
 |
Component:353; Jaccard:0.24488
 |
Component:353; Jaccard:0.25302
 |
Component:089; Jaccard:0.24406
 |
Correlation:-0.19678
  |
Correlation:0.34523
  |
Correlation:0.26442
  |
Correlation:0.26442
  |
Correlation:0.26442
  |
Correlation:0.26442
  |
Correlation:0.57654
  |
Component:089; Jaccard:0.12051
 |
Component:353; Jaccard:0.15503
 |
Component:069; Jaccard:0.18464
 |
Component:069; Jaccard:0.20919
 |
Component:069; Jaccard:0.21855
 |
Component:069; Jaccard:0.21146
 |
Component:069; Jaccard:0.19179
 |
Correlation:0.57654
  |
Correlation:0.26442
  |
Correlation:-0.19678
  |
Correlation:-0.19678
  |
Correlation:-0.19678
  |
Correlation:-0.19678
  |
Correlation:-0.19678
  |
Component:353; Jaccard:0.11792
 |
Component:069; Jaccard:0.15255
 |
Component:205; Jaccard:0.18393
 |
Component:205; Jaccard:0.19776
 |
Component:205; Jaccard:0.19415
 |
Component:205; Jaccard:0.18446
 |
Component:205; Jaccard:0.16608
 |
Correlation:0.26442
  |
Correlation:-0.19678
  |
Correlation:0.34523
  |
Correlation:0.34523
  |
Correlation:0.34523
  |
Correlation:0.34523
  |
Correlation:0.34523
  |
Component:037; Jaccard:0.1154
 |
Component:037; Jaccard:0.13273
 |
Component:104; Jaccard:0.15113
 |
Component:104; Jaccard:0.16203
 |
Component:104; Jaccard:0.15937
 |
Component:104; Jaccard:0.14721
 |
Component:243; Jaccard:0.12929
 |
Correlation:-0.059455
  |
Correlation:-0.059455
  |
Correlation:0.010949
  |
Correlation:0.010949
  |
Correlation:0.010949
  |
Correlation:0.010949
  |
Correlation:-0.094587
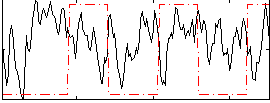  |
Component:266; Jaccard:0.11222
 |
Component:266; Jaccard:0.13052
 |
Component:377; Jaccard:0.14856
 |
Component:377; Jaccard:0.15856
 |
Component:243; Jaccard:0.15835
 |
Component:377; Jaccard:0.14703
 |
Component:377; Jaccard:0.12824
 |
Correlation:0.23534
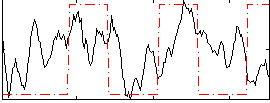  |
Correlation:0.23534
  |
Correlation:-0.16731
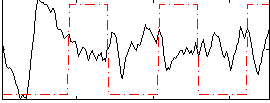  |
Correlation:-0.16731
  |
Correlation:-0.094587
  |
Correlation:-0.16731
  |
Correlation:-0.16731
  |
Component:243; Jaccard:0.10823
 |
Component:104; Jaccard:0.12953
 |
Component:243; Jaccard:0.14577
 |
Component:243; Jaccard:0.15588
 |
Component:377; Jaccard:0.15794
 |
Component:243; Jaccard:0.14596
 |
Component:104; Jaccard:0.12804
 |
Correlation:-0.094587
  |
Correlation:0.010949
  |
Correlation:-0.094587
  |
Correlation:-0.094587
  |
Correlation:-0.16731
  |
Correlation:-0.094587
  |
Correlation:0.010949
  |
Component:377; Jaccard:0.10752
 |
Component:377; Jaccard:0.12927
 |
Component:037; Jaccard:0.14481
 |
Component:037; Jaccard:0.14199
 |
Component:037; Jaccard:0.12856
 |
Component:037; Jaccard:0.10888
 |
Component:389; Jaccard:0.09968
 |
Correlation:-0.16731
  |
Correlation:-0.16731
  |
Correlation:-0.059455
  |
Correlation:-0.059455
  |
Correlation:-0.059455
  |
Correlation:-0.059455
  |
Correlation:0.18755
  |
Component:104; Jaccard:0.10738
 |
Component:243; Jaccard:0.12755
 |
Component:266; Jaccard:0.14349
 |
Component:266; Jaccard:0.13851
 |
Component:266; Jaccard:0.12342
 |
Component:389; Jaccard:0.10702
 |
Component:146; Jaccard:0.089582
 |
Correlation:0.010949
  |
Correlation:-0.094587
  |
Correlation:0.23534
  |
Correlation:0.23534
  |
Correlation:0.23534
  |
Correlation:0.18755
  |
Correlation:-0.1598
 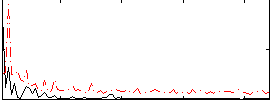 |
Component:376; Jaccard:0.10436
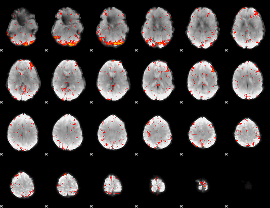 |
Component:376; Jaccard:0.115
 |
Component:146; Jaccard:0.1227
 |
Component:146; Jaccard:0.12609
 |
Component:146; Jaccard:0.11915
 |
Component:146; Jaccard:0.10626
 |
Component:037; Jaccard:0.087879
 |
Correlation:0.14437
  |
Correlation:0.14437
  |
Correlation:-0.1598
  |
Correlation:-0.1598
  |
Correlation:-0.1598
  |
Correlation:-0.1598
  |
Correlation:-0.059455
  |
Cope: 02:"SHAPES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
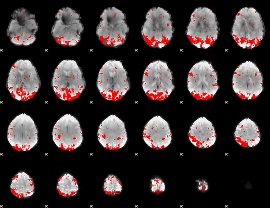 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
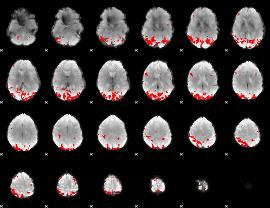 |
GLM binary result thresholded by 4
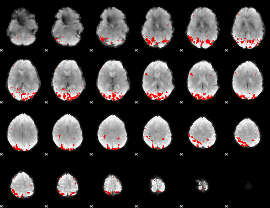 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
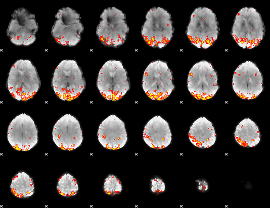 |
GLM Z-score result thresholded by 3.5
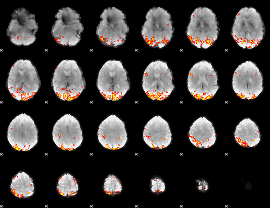 |
GLM Z-score result thresholded by 4
 |
Component:069; Jaccard:0.1423
 |
Component:069; Jaccard:0.18973
 |
Component:069; Jaccard:0.24406
 |
Component:069; Jaccard:0.2946
 |
Component:069; Jaccard:0.33101
 |
Component:069; Jaccard:0.34354
 |
Component:069; Jaccard:0.33226
 |
Correlation:0.37432
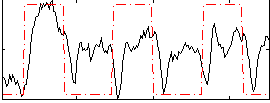  |
Correlation:0.37432
  |
Correlation:0.37432
  |
Correlation:0.37432
  |
Correlation:0.37432
  |
Correlation:0.37432
  |
Correlation:0.37432
  |
Component:037; Jaccard:0.13508
 |
Component:377; Jaccard:0.176
 |
Component:377; Jaccard:0.22263
 |
Component:377; Jaccard:0.25726
 |
Component:377; Jaccard:0.27266
 |
Component:377; Jaccard:0.26884
 |
Component:377; Jaccard:0.25572
 |
Correlation:0.19039
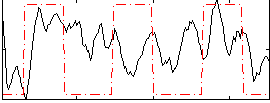  |
Correlation:0.3323
  |
Correlation:0.3323
  |
Correlation:0.3323
  |
Correlation:0.3323
  |
Correlation:0.3323
  |
Correlation:0.3323
  |
Component:377; Jaccard:0.13451
 |
Component:037; Jaccard:0.16624
 |
Component:037; Jaccard:0.188
 |
Component:332; Jaccard:0.20819
 |
Component:332; Jaccard:0.22371
 |
Component:332; Jaccard:0.22403
 |
Component:332; Jaccard:0.21007
 |
Correlation:0.3323
  |
Correlation:0.19039
  |
Correlation:0.19039
  |
Correlation:0.44855
  |
Correlation:0.44855
  |
Correlation:0.44855
  |
Correlation:0.44855
  |
Component:205; Jaccard:0.11964
 |
Component:332; Jaccard:0.14891
 |
Component:332; Jaccard:0.18128
 |
Component:037; Jaccard:0.20016
 |
Component:037; Jaccard:0.1944
 |
Component:037; Jaccard:0.17631
 |
Component:353; Jaccard:0.16912
 |
Correlation:-0.13233
  |
Correlation:0.44855
  |
Correlation:0.44855
  |
Correlation:0.19039
  |
Correlation:0.19039
  |
Correlation:0.19039
  |
Correlation:-0.20915
  |
Component:332; Jaccard:0.11725
 |
Component:205; Jaccard:0.14106
 |
Component:146; Jaccard:0.16096
 |
Component:146; Jaccard:0.17533
 |
Component:146; Jaccard:0.17779
 |
Component:353; Jaccard:0.17449
 |
Component:146; Jaccard:0.1599
 |
Correlation:0.44855
  |
Correlation:-0.13233
  |
Correlation:0.27154
  |
Correlation:0.27154
  |
Correlation:0.27154
  |
Correlation:-0.20915
  |
Correlation:0.27154
  |
Component:146; Jaccard:0.11572
 |
Component:146; Jaccard:0.13957
 |
Component:243; Jaccard:0.1593
 |
Component:243; Jaccard:0.17406
 |
Component:243; Jaccard:0.17718
 |
Component:146; Jaccard:0.17351
 |
Component:243; Jaccard:0.15568
 |
Correlation:0.27154
  |
Correlation:0.27154
  |
Correlation:0.13641
  |
Correlation:0.13641
  |
Correlation:0.13641
  |
Correlation:0.27154
  |
Correlation:0.13641
  |
Component:333; Jaccard:0.11536
 |
Component:243; Jaccard:0.13836
 |
Component:205; Jaccard:0.15804
 |
Component:205; Jaccard:0.16722
 |
Component:353; Jaccard:0.1718
 |
Component:243; Jaccard:0.17151
 |
Component:037; Jaccard:0.15175
 |
Correlation:0.17707
  |
Correlation:0.13641
  |
Correlation:-0.13233
  |
Correlation:-0.13233
  |
Correlation:-0.20915
  |
Correlation:0.13641
  |
Correlation:0.19039
  |
Component:288; Jaccard:0.11449
 |
Component:333; Jaccard:0.13093
 |
Component:104; Jaccard:0.14923
 |
Component:104; Jaccard:0.15942
 |
Component:205; Jaccard:0.16696
 |
Component:205; Jaccard:0.1604
 |
Component:205; Jaccard:0.14697
 |
Correlation:0.31311
  |
Correlation:0.17707
  |
Correlation:0.1848
  |
Correlation:0.1848
  |
Correlation:-0.13233
  |
Correlation:-0.13233
  |
Correlation:-0.13233
  |
Component:243; Jaccard:0.11445
 |
Component:104; Jaccard:0.12953
 |
Component:353; Jaccard:0.1445
 |
Component:353; Jaccard:0.15877
 |
Component:104; Jaccard:0.15859
 |
Component:104; Jaccard:0.14446
 |
Component:104; Jaccard:0.13034
 |
Correlation:0.13641
  |
Correlation:0.1848
  |
Correlation:-0.20915
  |
Correlation:-0.20915
  |
Correlation:0.1848
  |
Correlation:0.1848
  |
Correlation:0.1848
  |
Component:218; Jaccard:0.10811
 |
Component:288; Jaccard:0.12667
 |
Component:333; Jaccard:0.13998
 |
Component:333; Jaccard:0.13643
 |
Component:333; Jaccard:0.12155
 |
Component:020; Jaccard:0.10526
 |
Component:020; Jaccard:0.092208
 |
Correlation:0.14573
  |
Correlation:0.31311
  |
Correlation:0.17707
  |
Correlation:0.17707
  |
Correlation:0.17707
  |
Correlation:0.2781
  |
Correlation:0.2781
  |
Cope: 03:"FACES-SHAPES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
GLM binary result thresholded by 4
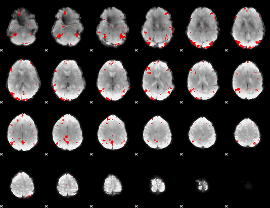 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
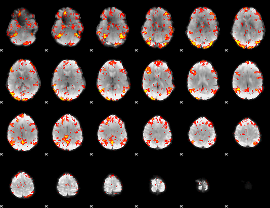 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
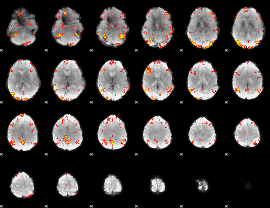 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:032; Jaccard:0.13095
 |
Component:032; Jaccard:0.16647
 |
Component:032; Jaccard:0.2022
 |
Component:089; Jaccard:0.22821
 |
Component:089; Jaccard:0.25316
 |
Component:089; Jaccard:0.27021
 |
Component:089; Jaccard:0.26491
 |
Correlation:0.38203
  |
Correlation:0.38203
  |
Correlation:0.38203
  |
Correlation:0.56809
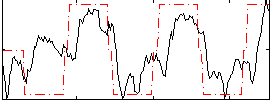 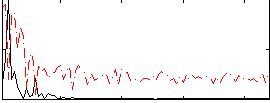 |
Correlation:0.56809
  |
Correlation:0.56809
  |
Correlation:0.56809
  |
Component:077; Jaccard:0.12515
 |
Component:077; Jaccard:0.15457
 |
Component:089; Jaccard:0.19185
 |
Component:032; Jaccard:0.2263
 |
Component:032; Jaccard:0.22999
 |
Component:033; Jaccard:0.21323
 |
Component:033; Jaccard:0.19412
 |
Correlation:0.47928
  |
Correlation:0.47928
  |
Correlation:0.56809
  |
Correlation:0.38203
  |
Correlation:0.38203
  |
Correlation:0.60182
  |
Correlation:0.60182
  |
Component:089; Jaccard:0.1207
 |
Component:089; Jaccard:0.15368
 |
Component:077; Jaccard:0.18447
 |
Component:077; Jaccard:0.20595
 |
Component:077; Jaccard:0.21275
 |
Component:032; Jaccard:0.2111
 |
Component:077; Jaccard:0.18378
 |
Correlation:0.56809
  |
Correlation:0.56809
  |
Correlation:0.47928
  |
Correlation:0.47928
  |
Correlation:0.47928
  |
Correlation:0.38203
  |
Correlation:0.47928
  |
Component:315; Jaccard:0.11988
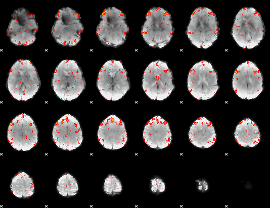 |
Component:033; Jaccard:0.13927
 |
Component:033; Jaccard:0.17
 |
Component:033; Jaccard:0.19833
 |
Component:033; Jaccard:0.21257
 |
Component:077; Jaccard:0.20295
 |
Component:032; Jaccard:0.18145
 |
Correlation:0.21767
  |
Correlation:0.60182
  |
Correlation:0.60182
  |
Correlation:0.60182
  |
Correlation:0.60182
  |
Correlation:0.47928
  |
Correlation:0.38203
  |
Component:355; Jaccard:0.11894
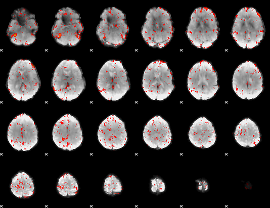 |
Component:355; Jaccard:0.13796
 |
Component:355; Jaccard:0.15308
 |
Component:355; Jaccard:0.15889
 |
Component:355; Jaccard:0.15479
 |
Component:355; Jaccard:0.14011
 |
Component:355; Jaccard:0.11957
 |
Correlation:0.25196
  |
Correlation:0.25196
  |
Correlation:0.25196
  |
Correlation:0.25196
  |
Correlation:0.25196
  |
Correlation:0.25196
  |
Correlation:0.25196
  |
Component:266; Jaccard:0.11277
 |
Component:315; Jaccard:0.13746
 |
Component:315; Jaccard:0.14683
 |
Component:315; Jaccard:0.14401
 |
Component:266; Jaccard:0.13195
 |
Component:389; Jaccard:0.118
 |
Component:389; Jaccard:0.11275
 |
Correlation:0.19072
 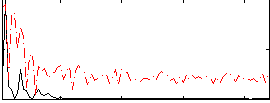 |
Correlation:0.21767
  |
Correlation:0.21767
  |
Correlation:0.21767
  |
Correlation:0.19072
  |
Correlation:0.19191
  |
Correlation:0.19191
  |
Component:033; Jaccard:0.11118
 |
Component:266; Jaccard:0.12765
 |
Component:266; Jaccard:0.13996
 |
Component:266; Jaccard:0.14198
 |
Component:315; Jaccard:0.12669
 |
Component:266; Jaccard:0.11163
 |
Component:353; Jaccard:0.096718
 |
Correlation:0.60182
  |
Correlation:0.19072
  |
Correlation:0.19072
  |
Correlation:0.19072
  |
Correlation:0.21767
  |
Correlation:0.19072
  |
Correlation:0.25687
  |
Component:129; Jaccard:0.09054
 |
Component:159; Jaccard:0.097057
 |
Component:389; Jaccard:0.10275
 |
Component:389; Jaccard:0.11106
 |
Component:389; Jaccard:0.11879
 |
Component:353; Jaccard:0.10336
 |
Component:266; Jaccard:0.088172
 |
Correlation:-0.00178
  |
Correlation:0.25314
  |
Correlation:0.19191
  |
Correlation:0.19191
  |
Correlation:0.19191
  |
Correlation:0.25687
  |
Correlation:0.19072
  |
Component:159; Jaccard:0.090241
 |
Component:129; Jaccard:0.096435
 |
Component:270; Jaccard:0.10168
 |
Component:270; Jaccard:0.10592
 |
Component:270; Jaccard:0.10327
 |
Component:315; Jaccard:0.10119
 |
Component:270; Jaccard:0.078505
 |
Correlation:0.25314
  |
Correlation:-0.00178
  |
Correlation:0.38944
  |
Correlation:0.38944
  |
Correlation:0.38944
  |
Correlation:0.21767
  |
Correlation:0.38944
  |
Component:376; Jaccard:0.087631
 |
Component:376; Jaccard:0.095332
 |
Component:376; Jaccard:0.098372
 |
Component:353; Jaccard:0.097847
 |
Component:353; Jaccard:0.10254
 |
Component:270; Jaccard:0.09328
 |
Component:315; Jaccard:0.074012
 |
Correlation:0.081485
  |
Correlation:0.081485
  |
Correlation:0.081485
  |
Correlation:0.25687
  |
Correlation:0.25687
  |
Correlation:0.38944
  |
Correlation:0.21767
  |
Cope: 04:"neg_FACES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
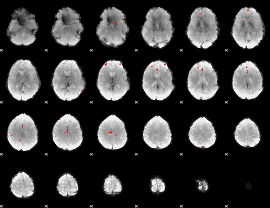 |
GLM binary result thresholded by 4
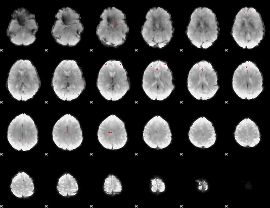 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:341; Jaccard:0.1214
 |
Component:341; Jaccard:0.14317
 |
Component:341; Jaccard:0.14532
 |
Component:341; Jaccard:0.11897
 |
Component:341; Jaccard:0.075896
 |
Component:341; Jaccard:0.041794
 |
Component:341; Jaccard:0.021307
 |
Correlation:0.27783
  |
Correlation:0.27783
  |
Correlation:0.27783
  |
Correlation:0.27783
  |
Correlation:0.27783
  |
Correlation:0.27783
  |
Correlation:0.27783
  |
Component:115; Jaccard:0.11657
 |
Component:115; Jaccard:0.12949
 |
Component:115; Jaccard:0.12339
 |
Component:115; Jaccard:0.092696
 |
Component:115; Jaccard:0.053471
 |
Component:115; Jaccard:0.02929
 |
Component:115; Jaccard:0.015355
 |
Correlation:-0.003566
  |
Correlation:-0.003566
  |
Correlation:-0.003566
  |
Correlation:-0.003566
  |
Correlation:-0.003566
  |
Correlation:-0.003566
  |
Correlation:-0.003566
  |
Component:258; Jaccard:0.082231
 |
Component:258; Jaccard:0.089412
 |
Component:258; Jaccard:0.085111
 |
Component:121; Jaccard:0.06496
 |
Component:121; Jaccard:0.042021
 |
Component:026; Jaccard:0.025556
 |
Component:026; Jaccard:0.012789
 |
Correlation:0.27735
  |
Correlation:0.27735
  |
Correlation:0.27735
  |
Correlation:0.16391
  |
Correlation:0.16391
  |
Correlation:0.32937
  |
Correlation:0.32937
  |
Component:026; Jaccard:0.079282
 |
Component:121; Jaccard:0.08457
 |
Component:121; Jaccard:0.080608
 |
Component:258; Jaccard:0.064714
 |
Component:026; Jaccard:0.041587
 |
Component:121; Jaccard:0.0234
 |
Component:258; Jaccard:0.011553
 |
Correlation:0.32937
  |
Correlation:0.16391
  |
Correlation:0.16391
  |
Correlation:0.27735
  |
Correlation:0.32937
  |
Correlation:0.16391
  |
Correlation:0.27735
  |
Component:257; Jaccard:0.078166
 |
Component:026; Jaccard:0.083824
 |
Component:026; Jaccard:0.080393
 |
Component:026; Jaccard:0.063785
 |
Component:258; Jaccard:0.039461
 |
Component:258; Jaccard:0.022712
 |
Component:121; Jaccard:0.011291
 |
Correlation:0.089675
 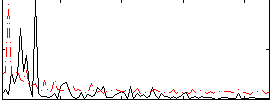 |
Correlation:0.32937
  |
Correlation:0.32937
  |
Correlation:0.32937
  |
Correlation:0.27735
  |
Correlation:0.27735
  |
Correlation:0.16391
  |
Component:299; Jaccard:0.078039
 |
Component:299; Jaccard:0.083454
 |
Component:299; Jaccard:0.077319
 |
Component:299; Jaccard:0.060546
 |
Component:299; Jaccard:0.03907
 |
Component:299; Jaccard:0.021593
 |
Component:299; Jaccard:0.009217
 |
Correlation:-0.068592
 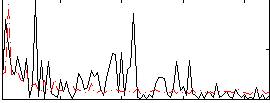 |
Correlation:-0.068592
  |
Correlation:-0.068592
  |
Correlation:-0.068592
  |
Correlation:-0.068592
  |
Correlation:-0.068592
  |
Correlation:-0.068592
  |
Component:121; Jaccard:0.076953
 |
Component:257; Jaccard:0.076157
 |
Component:155; Jaccard:0.067
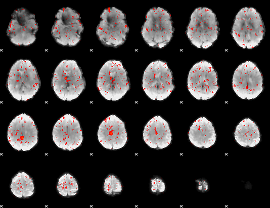 |
Component:155; Jaccard:0.05043
 |
Component:155; Jaccard:0.029315
 |
Component:155; Jaccard:0.016169
 |
Component:155; Jaccard:0.008306
 |
Correlation:0.16391
  |
Correlation:0.089675
  |
Correlation:-0.092356
  |
Correlation:-0.092356
  |
Correlation:-0.092356
  |
Correlation:-0.092356
  |
Correlation:-0.092356
  |
Component:399; Jaccard:0.071668
 |
Component:155; Jaccard:0.072238
 |
Component:257; Jaccard:0.064514
 |
Component:008; Jaccard:0.04498
 |
Component:008; Jaccard:0.026823
 |
Component:008; Jaccard:0.012059
 |
Component:111; Jaccard:0.006886
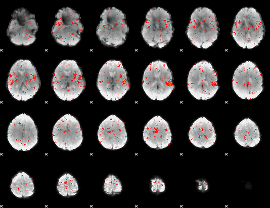 |
Correlation:-0.12532
  |
Correlation:-0.092356
  |
Correlation:0.089675
  |
Correlation:0.16319
  |
Correlation:0.16319
  |
Correlation:0.16319
  |
Correlation:0.12477
 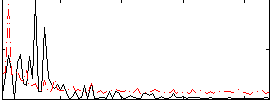 |
Component:208; Jaccard:0.071287
 |
Component:208; Jaccard:0.06834
 |
Component:396; Jaccard:0.060415
 |
Component:396; Jaccard:0.044672
 |
Component:396; Jaccard:0.025293
 |
Component:396; Jaccard:0.01203
 |
Component:396; Jaccard:0.006522
 |
Correlation:-0.15504
  |
Correlation:-0.15504
  |
Correlation:0.42104
  |
Correlation:0.42104
  |
Correlation:0.42104
  |
Correlation:0.42104
  |
Correlation:0.42104
  |
Component:155; Jaccard:0.07104
 |
Component:356; Jaccard:0.067129
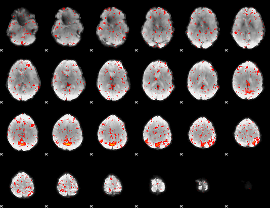 |
Component:208; Jaccard:0.058537
 |
Component:255; Jaccard:0.042722
 |
Component:255; Jaccard:0.023112
 |
Component:255; Jaccard:0.011811
 |
Component:269; Jaccard:0.006363
 |
Correlation:-0.092356
  |
Correlation:-0.11963
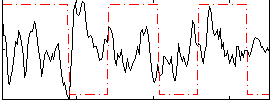 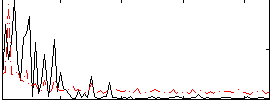 |
Correlation:-0.15504
  |
Correlation:0.14878
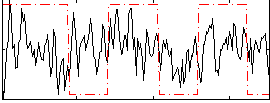  |
Correlation:0.14878
  |
Correlation:0.14878
  |
Correlation:0.17471
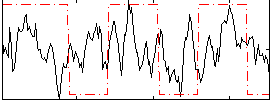  |
Cope: 05:"neg_SHAPES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
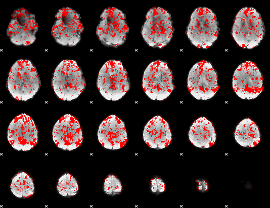 |
GLM binary result thresholded by 1.5
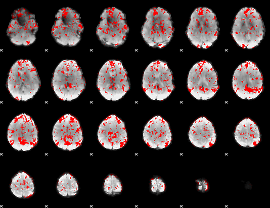 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
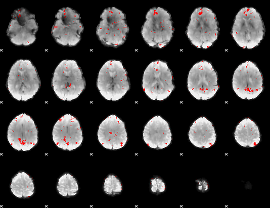 |
GLM binary result thresholded by 3.5
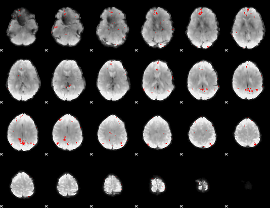 |
GLM binary result thresholded by 4
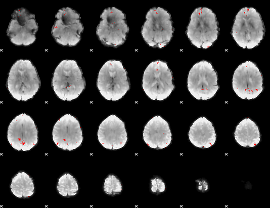 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
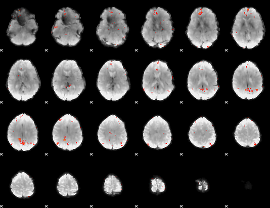 |
GLM Z-score result thresholded by 4
 |
Component:159; Jaccard:0.13197
 |
Component:159; Jaccard:0.16093
 |
Component:159; Jaccard:0.17955
 |
Component:159; Jaccard:0.16912
 |
Component:159; Jaccard:0.12911
 |
Component:159; Jaccard:0.0866
 |
Component:159; Jaccard:0.048787
 |
Correlation:0.31739
  |
Correlation:0.31739
  |
Correlation:0.31739
  |
Correlation:0.31739
  |
Correlation:0.31739
  |
Correlation:0.31739
  |
Correlation:0.31739
  |
Component:077; Jaccard:0.10351
 |
Component:077; Jaccard:0.11766
 |
Component:077; Jaccard:0.12022
 |
Component:270; Jaccard:0.11606
 |
Component:270; Jaccard:0.09324
 |
Component:270; Jaccard:0.061381
 |
Component:270; Jaccard:0.036766
 |
Correlation:0.47476
  |
Correlation:0.47476
  |
Correlation:0.47476
  |
Correlation:0.37966
  |
Correlation:0.37966
  |
Correlation:0.37966
  |
Correlation:0.37966
  |
Component:115; Jaccard:0.099878
 |
Component:270; Jaccard:0.10626
 |
Component:270; Jaccard:0.1182
 |
Component:077; Jaccard:0.11298
 |
Component:077; Jaccard:0.089478
 |
Component:077; Jaccard:0.059424
 |
Component:077; Jaccard:0.036435
 |
Correlation:0.02278
  |
Correlation:0.37966
  |
Correlation:0.37966
  |
Correlation:0.47476
  |
Correlation:0.47476
  |
Correlation:0.47476
  |
Correlation:0.47476
  |
Component:270; Jaccard:0.08829
 |
Component:115; Jaccard:0.10536
 |
Component:115; Jaccard:0.1034
 |
Component:235; Jaccard:0.090283
 |
Component:235; Jaccard:0.068537
 |
Component:235; Jaccard:0.04267
 |
Component:235; Jaccard:0.021813
 |
Correlation:0.37966
  |
Correlation:0.02278
  |
Correlation:0.02278
  |
Correlation:0.17953
  |
Correlation:0.17953
  |
Correlation:0.17953
  |
Correlation:0.17953
  |
Component:235; Jaccard:0.087419
 |
Component:235; Jaccard:0.098021
 |
Component:235; Jaccard:0.10115
 |
Component:115; Jaccard:0.083453
 |
Component:072; Jaccard:0.061296
 |
Component:072; Jaccard:0.037479
 |
Component:072; Jaccard:0.020484
 |
Correlation:0.17953
  |
Correlation:0.17953
  |
Correlation:0.17953
  |
Correlation:0.02278
  |
Correlation:0.26615
  |
Correlation:0.26615
  |
Correlation:0.26615
  |
Component:032; Jaccard:0.082334
 |
Component:032; Jaccard:0.08862
 |
Component:072; Jaccard:0.090956
 |
Component:072; Jaccard:0.083196
 |
Component:115; Jaccard:0.056047
 |
Component:035; Jaccard:0.033474
 |
Component:186; Jaccard:0.019123
 |
Correlation:0.31848
  |
Correlation:0.31848
  |
Correlation:0.26615
  |
Correlation:0.26615
  |
Correlation:0.02278
  |
Correlation:0.37546
  |
Correlation:0.019022
  |
Component:356; Jaccard:0.082096
 |
Component:035; Jaccard:0.085063
 |
Component:032; Jaccard:0.086135
 |
Component:035; Jaccard:0.072161
 |
Component:035; Jaccard:0.053699
 |
Component:115; Jaccard:0.032364
 |
Component:035; Jaccard:0.018497
 |
Correlation:0.020344
  |
Correlation:0.37546
  |
Correlation:0.31848
  |
Correlation:0.37546
  |
Correlation:0.37546
  |
Correlation:0.02278
  |
Correlation:0.37546
  |
Component:229; Jaccard:0.081618
 |
Component:072; Jaccard:0.085038
 |
Component:035; Jaccard:0.084692
 |
Component:032; Jaccard:0.071386
 |
Component:032; Jaccard:0.050945
 |
Component:229; Jaccard:0.030631
 |
Component:229; Jaccard:0.018314
 |
Correlation:0.084935
  |
Correlation:0.26615
  |
Correlation:0.37546
  |
Correlation:0.31848
  |
Correlation:0.31848
  |
Correlation:0.084935
  |
Correlation:0.084935
  |
Component:035; Jaccard:0.08107
 |
Component:229; Jaccard:0.08355
 |
Component:229; Jaccard:0.077614
 |
Component:229; Jaccard:0.065202
 |
Component:229; Jaccard:0.046467
 |
Component:186; Jaccard:0.03027
 |
Component:032; Jaccard:0.016255
 |
Correlation:0.37546
  |
Correlation:0.084935
  |
Correlation:0.084935
  |
Correlation:0.084935
  |
Correlation:0.084935
  |
Correlation:0.019022
  |
Correlation:0.31848
  |
Component:399; Jaccard:0.08007
 |
Component:356; Jaccard:0.080122
 |
Component:356; Jaccard:0.071448
 |
Component:299; Jaccard:0.062022
 |
Component:299; Jaccard:0.045702
 |
Component:032; Jaccard:0.029363
 |
Component:375; Jaccard:0.015028
 |
Correlation:0.11896
  |
Correlation:0.020344
  |
Correlation:0.020344
  |
Correlation:0.11629
  |
Correlation:0.11629
  |
Correlation:0.31848
  |
Correlation:0.056938
  |
Cope: 06:"SHAPES-FACES" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
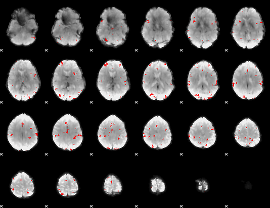
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
 |
GLM binary result thresholded by 3.5
 |
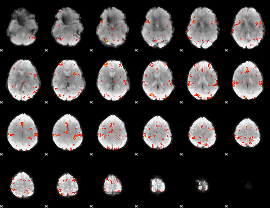
GLM binary result thresholded by 4
 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
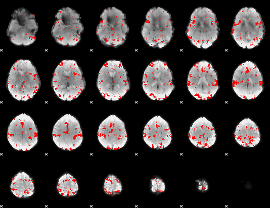
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:026; Jaccard:0.11986
 |
Component:026; Jaccard:0.14618
 |
Component:026; Jaccard:0.16895
 |
Component:258; Jaccard:0.18069
 |
Component:026; Jaccard:0.17081
 |
Component:258; Jaccard:0.14632
 |
Component:258; Jaccard:0.1092
 |
Correlation:0.36249
  |
Correlation:0.36249
  |
Correlation:0.36249
  |
Correlation:0.37123
  |
Correlation:0.36249
  |
Correlation:0.37123
  |
Correlation:0.37123
  |
Component:332; Jaccard:0.11482
 |
Component:258; Jaccard:0.14073
 |
Component:258; Jaccard:0.16769
 |
Component:026; Jaccard:0.17933
 |
Component:258; Jaccard:0.17044
 |
Component:026; Jaccard:0.14553
 |
Component:026; Jaccard:0.10698
 |
Correlation:0.51558
  |
Correlation:0.37123
  |
Correlation:0.37123
  |
Correlation:0.36249
  |
Correlation:0.37123
  |
Correlation:0.36249
  |
Correlation:0.36249
  |
Component:258; Jaccard:0.11335
 |
Component:332; Jaccard:0.13969
 |
Component:332; Jaccard:0.1661
 |
Component:332; Jaccard:0.17851
 |
Component:332; Jaccard:0.16653
 |
Component:332; Jaccard:0.13937
 |
Component:332; Jaccard:0.10171
 |
Correlation:0.37123
  |
Correlation:0.51558
  |
Correlation:0.51558
  |
Correlation:0.51558
  |
Correlation:0.51558
  |
Correlation:0.51558
  |
Correlation:0.51558
  |
Component:325; Jaccard:0.10816
 |
Component:396; Jaccard:0.13001
 |
Component:396; Jaccard:0.14976
 |
Component:396; Jaccard:0.15879
 |
Component:396; Jaccard:0.14759
 |
Component:396; Jaccard:0.12119
 |
Component:396; Jaccard:0.083636
 |
Correlation:0.3566
  |
Correlation:0.47207
  |
Correlation:0.47207
  |
Correlation:0.47207
  |
Correlation:0.47207
  |
Correlation:0.47207
  |
Correlation:0.47207
  |
Component:396; Jaccard:0.10714
 |
Component:325; Jaccard:0.12253
 |
Component:325; Jaccard:0.13159
 |
Component:325; Jaccard:0.12937
 |
Component:325; Jaccard:0.11185
 |
Component:325; Jaccard:0.086245
 |
Component:377; Jaccard:0.060869
 |
Correlation:0.47207
  |
Correlation:0.3566
  |
Correlation:0.3566
  |
Correlation:0.3566
  |
Correlation:0.3566
  |
Correlation:0.3566
  |
Correlation:0.271
  |
Component:020; Jaccard:0.10088
 |
Component:020; Jaccard:0.1134
 |
Component:020; Jaccard:0.12168
 |
Component:020; Jaccard:0.1209
 |
Component:020; Jaccard:0.10853
 |
Component:020; Jaccard:0.085606
 |
Component:020; Jaccard:0.060377
 |
Correlation:0.21439
  |
Correlation:0.21439
  |
Correlation:0.21439
  |
Correlation:0.21439
  |
Correlation:0.21439
  |
Correlation:0.21439
  |
Correlation:0.21439
  |
Component:013; Jaccard:0.096558
 |
Component:013; Jaccard:0.10994
 |
Component:013; Jaccard:0.11803
 |
Component:377; Jaccard:0.11266
 |
Component:377; Jaccard:0.1013
 |
Component:377; Jaccard:0.082814
 |
Component:325; Jaccard:0.058829
 |
Correlation:0.27103
  |
Correlation:0.27103
  |
Correlation:0.27103
  |
Correlation:0.271
  |
Correlation:0.271
  |
Correlation:0.271
  |
Correlation:0.3566
  |
Component:377; Jaccard:0.094776
 |
Component:377; Jaccard:0.10757
 |
Component:377; Jaccard:0.11567
 |
Component:013; Jaccard:0.11139
 |
Component:343; Jaccard:0.096789
 |
Component:343; Jaccard:0.078737
 |
Component:343; Jaccard:0.056899
 |
Correlation:0.271
  |
Correlation:0.271
  |
Correlation:0.271
  |
Correlation:0.27103
  |
Correlation:0.26816
  |
Correlation:0.26816
  |
Correlation:0.26816
  |
Component:382; Jaccard:0.093597
 |
Component:080; Jaccard:0.10073
 |
Component:343; Jaccard:0.10764
 |
Component:080; Jaccard:0.10798
 |
Component:069; Jaccard:0.096465
 |
Component:069; Jaccard:0.077982
 |
Component:069; Jaccard:0.056276
 |
Correlation:0.19292
  |
Correlation:0.3107
  |
Correlation:0.26816
  |
Correlation:0.3107
  |
Correlation:0.30977
  |
Correlation:0.30977
  |
Correlation:0.30977
  |
Component:288; Jaccard:0.091704
 |
Component:343; Jaccard:0.098337
 |
Component:080; Jaccard:0.10733
 |
Component:069; Jaccard:0.10678
 |
Component:013; Jaccard:0.095596
 |
Component:013; Jaccard:0.071776
 |
Component:063; Jaccard:0.049704
 |
Correlation:0.30298
  |
Correlation:0.26816
  |
Correlation:0.3107
  |
Correlation:0.30977
  |
Correlation:0.27103
  |
Correlation:0.27103
  |
Correlation:0.21952
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































