Task name: RELATIONAL, TR=0.72, totally 232 volumes, 10 waves, 6 copes BACK
|
Cope: 01:"MATCH" BACK
|
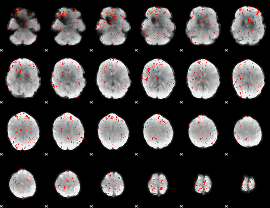
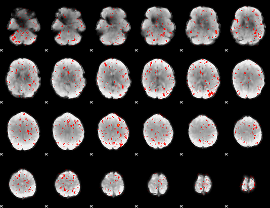
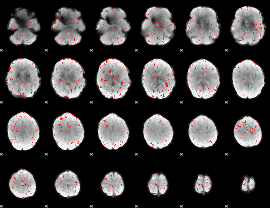
GLM result group-wise
 |
GLM binary result thresholded by 1
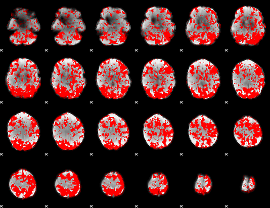 |
GLM binary result thresholded by 1.5
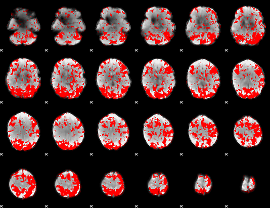 |
GLM binary result thresholded by 2
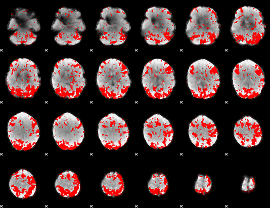 |
GLM binary result thresholded by 2.5
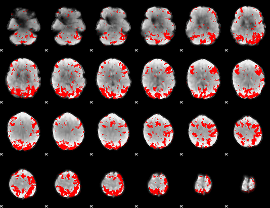 |
GLM binary result thresholded by 3
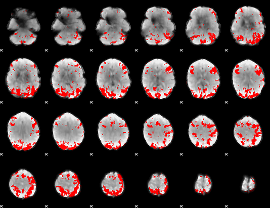 |
GLM binary result thresholded by 3.5
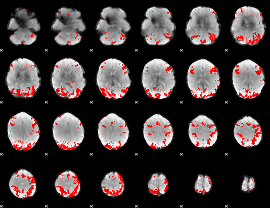 |
GLM binary result thresholded by 4
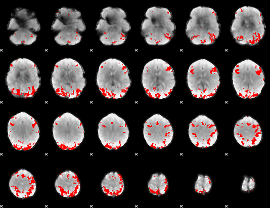 |
GLM Z-score result thresholded by 1
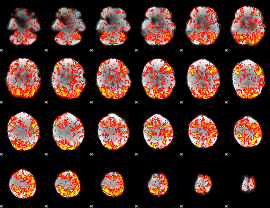 |
GLM Z-score result thresholded by 1.5
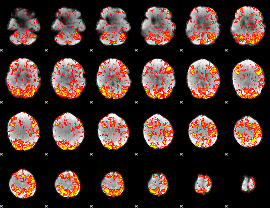 |
GLM Z-score result thresholded by 2
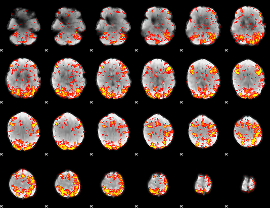 |
GLM Z-score result thresholded by 2.5
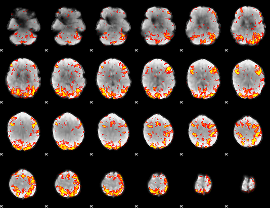 |
GLM Z-score result thresholded by 3
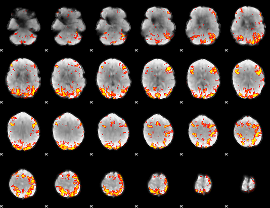 |
GLM Z-score result thresholded by 3.5
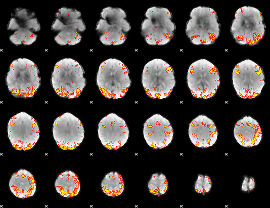 |
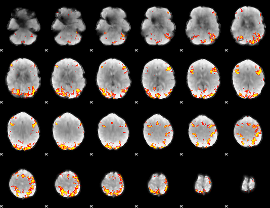
GLM Z-score result thresholded by 4
 |
Component:234; Jaccard:0.13023
 |
Component:234; Jaccard:0.16194
 |
Component:087; Jaccard:0.20187
 |
Component:087; Jaccard:0.24755
 |
Component:087; Jaccard:0.29022
 |
Component:087; Jaccard:0.32535
 |
Component:087; Jaccard:0.34972
 |
Correlation:0.16793
 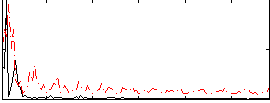 |
Correlation:0.16793
  |
Correlation:0.27178
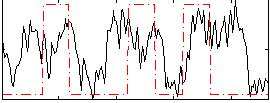 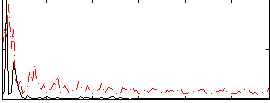 |
Correlation:0.27178
  |
Correlation:0.27178
  |
Correlation:0.27178
  |
Correlation:0.27178
  |
Component:087; Jaccard:0.12374
 |
Component:087; Jaccard:0.15861
 |
Component:234; Jaccard:0.19698
 |
Component:234; Jaccard:0.22902
 |
Component:040; Jaccard:0.26103
 |
Component:040; Jaccard:0.28477
 |
Component:040; Jaccard:0.29296
 |
Correlation:0.27178
  |
Correlation:0.27178
  |
Correlation:0.16793
  |
Correlation:0.16793
  |
Correlation:0.25127
 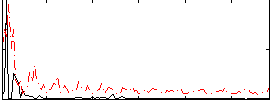 |
Correlation:0.25127
  |
Correlation:0.25127
  |
Component:257; Jaccard:0.11941
 |
Component:048; Jaccard:0.14624
 |
Component:040; Jaccard:0.18304
 |
Component:040; Jaccard:0.22363
 |
Component:234; Jaccard:0.25237
 |
Component:234; Jaccard:0.26348
 |
Component:048; Jaccard:0.25919
 |
Correlation:0.10634
 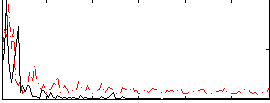 |
Correlation:0.15743
  |
Correlation:0.25127
  |
Correlation:0.25127
  |
Correlation:0.16793
  |
Correlation:0.16793
  |
Correlation:0.15743
  |
Component:048; Jaccard:0.11731
 |
Component:040; Jaccard:0.14386
 |
Component:048; Jaccard:0.17935
 |
Component:048; Jaccard:0.2106
 |
Component:048; Jaccard:0.23769
 |
Component:048; Jaccard:0.25331
 |
Component:234; Jaccard:0.2589
 |
Correlation:0.15743
  |
Correlation:0.25127
  |
Correlation:0.15743
  |
Correlation:0.15743
  |
Correlation:0.15743
  |
Correlation:0.15743
  |
Correlation:0.16793
  |
Component:299; Jaccard:0.11368
 |
Component:257; Jaccard:0.1416
 |
Component:257; Jaccard:0.16207
 |
Component:037; Jaccard:0.18091
 |
Component:037; Jaccard:0.20157
 |
Component:037; Jaccard:0.21414
 |
Component:037; Jaccard:0.22061
 |
Correlation:0.018923
  |
Correlation:0.10634
  |
Correlation:0.10634
  |
Correlation:0.10379
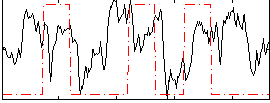 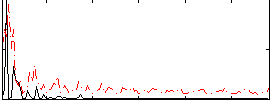 |
Correlation:0.10379
  |
Correlation:0.10379
  |
Correlation:0.10379
  |
Component:111; Jaccard:0.1124
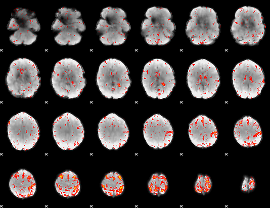 |
Component:299; Jaccard:0.1347
 |
Component:299; Jaccard:0.15611
 |
Component:257; Jaccard:0.1772
 |
Component:295; Jaccard:0.18933
 |
Component:295; Jaccard:0.19879
 |
Component:295; Jaccard:0.20175
 |
Correlation:-0.0055805
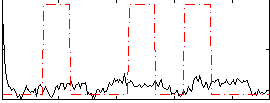 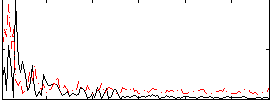 |
Correlation:0.018923
  |
Correlation:0.018923
  |
Correlation:0.10634
  |
Correlation:0.0761
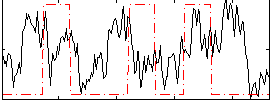 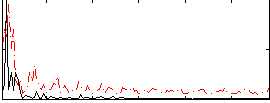 |
Correlation:0.0761
  |
Correlation:0.0761
  |
Component:040; Jaccard:0.11215
 |
Component:037; Jaccard:0.13013
 |
Component:037; Jaccard:0.15517
 |
Component:299; Jaccard:0.17288
 |
Component:257; Jaccard:0.18363
 |
Component:257; Jaccard:0.18208
 |
Component:299; Jaccard:0.17419
 |
Correlation:0.25127
  |
Correlation:0.10379
  |
Correlation:0.10379
  |
Correlation:0.018923
  |
Correlation:0.10634
  |
Correlation:0.10634
  |
Correlation:0.018923
  |
Component:400; Jaccard:0.10756
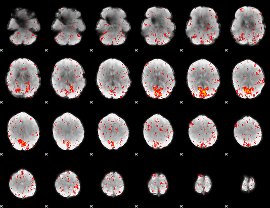 |
Component:295; Jaccard:0.1285
 |
Component:295; Jaccard:0.1518
 |
Component:295; Jaccard:0.17198
 |
Component:299; Jaccard:0.18034
 |
Component:299; Jaccard:0.18027
 |
Component:257; Jaccard:0.17181
 |
Correlation:0.029991
 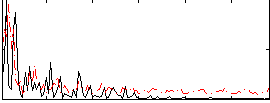 |
Correlation:0.0761
  |
Correlation:0.0761
  |
Correlation:0.0761
  |
Correlation:0.018923
  |
Correlation:0.018923
  |
Correlation:0.10634
  |
Component:037; Jaccard:0.10744
 |
Component:079; Jaccard:0.12333
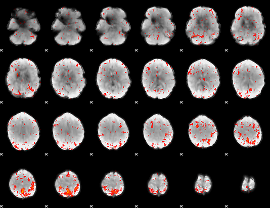 |
Component:079; Jaccard:0.14159
 |
Component:079; Jaccard:0.156
 |
Component:079; Jaccard:0.16228
 |
Component:079; Jaccard:0.1623
 |
Component:079; Jaccard:0.15573
 |
Correlation:0.10379
  |
Correlation:0.15221
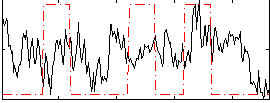 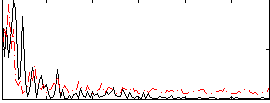 |
Correlation:0.15221
  |
Correlation:0.15221
  |
Correlation:0.15221
  |
Correlation:0.15221
  |
Correlation:0.15221
  |
Component:295; Jaccard:0.10732
 |
Component:111; Jaccard:0.12016
 |
Component:400; Jaccard:0.13025
 |
Component:367; Jaccard:0.14351
 |
Component:367; Jaccard:0.15258
 |
Component:367; Jaccard:0.15621
 |
Component:367; Jaccard:0.15523
 |
Correlation:0.0761
  |
Correlation:-0.0055805
  |
Correlation:0.029991
  |
Correlation:0.14643
  |
Correlation:0.14643
  |
Correlation:0.14643
  |
Correlation:0.14643
  |
Cope: 02:"REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
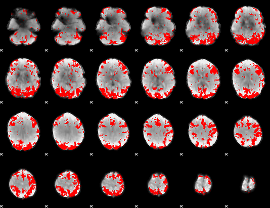 |
GLM binary result thresholded by 3.5
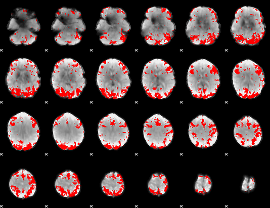 |
GLM binary result thresholded by 4
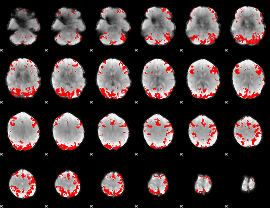 |
GLM Z-score result thresholded by 1
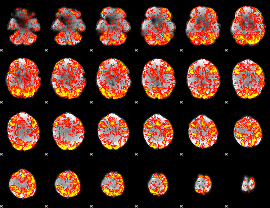 |
GLM Z-score result thresholded by 1.5
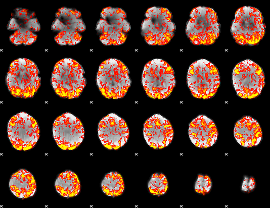 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
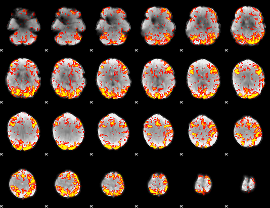 |
GLM Z-score result thresholded by 3
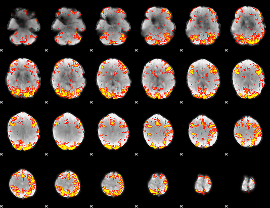 |
GLM Z-score result thresholded by 3.5
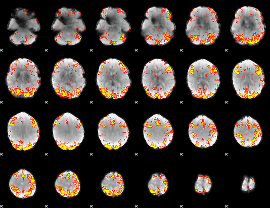 |
GLM Z-score result thresholded by 4
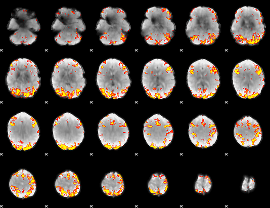 |
Component:257; Jaccard:0.1055
 |
Component:257; Jaccard:0.12502
 |
Component:087; Jaccard:0.15121
 |
Component:087; Jaccard:0.18473
 |
Component:087; Jaccard:0.21957
 |
Component:087; Jaccard:0.25437
 |
Component:087; Jaccard:0.28705
 |
Correlation:0.25634
  |
Correlation:0.25634
  |
Correlation:0.29962
  |
Correlation:0.29962
  |
Correlation:0.29962
  |
Correlation:0.29962
  |
Correlation:0.29962
  |
Component:234; Jaccard:0.10429
 |
Component:234; Jaccard:0.12412
 |
Component:257; Jaccard:0.14789
 |
Component:048; Jaccard:0.17509
 |
Component:048; Jaccard:0.20834
 |
Component:048; Jaccard:0.23768
 |
Component:048; Jaccard:0.26267
 |
Correlation:0.23244
  |
Correlation:0.23244
  |
Correlation:0.25634
  |
Correlation:0.22224
  |
Correlation:0.22224
  |
Correlation:0.22224
  |
Correlation:0.22224
  |
Component:087; Jaccard:0.10026
 |
Component:087; Jaccard:0.1225
 |
Component:234; Jaccard:0.14667
 |
Component:257; Jaccard:0.1714
 |
Component:295; Jaccard:0.19522
 |
Component:040; Jaccard:0.2215
 |
Component:040; Jaccard:0.24349
 |
Correlation:0.29962
  |
Correlation:0.29962
  |
Correlation:0.23244
  |
Correlation:0.25634
  |
Correlation:0.45138
  |
Correlation:0.27494
  |
Correlation:0.27494
  |
Component:295; Jaccard:0.098777
 |
Component:295; Jaccard:0.11888
 |
Component:048; Jaccard:0.14495
 |
Component:234; Jaccard:0.1713
 |
Component:037; Jaccard:0.19422
 |
Component:037; Jaccard:0.21942
 |
Component:037; Jaccard:0.24259
 |
Correlation:0.45138
  |
Correlation:0.45138
  |
Correlation:0.22224
  |
Correlation:0.23244
  |
Correlation:0.38412
 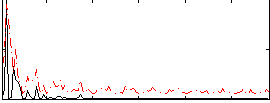 |
Correlation:0.38412
  |
Correlation:0.38412
  |
Component:048; Jaccard:0.097698
 |
Component:048; Jaccard:0.11873
 |
Component:295; Jaccard:0.14318
 |
Component:295; Jaccard:0.16894
 |
Component:234; Jaccard:0.19324
 |
Component:295; Jaccard:0.21637
 |
Component:295; Jaccard:0.23662
 |
Correlation:0.22224
  |
Correlation:0.22224
  |
Correlation:0.45138
  |
Correlation:0.45138
  |
Correlation:0.23244
  |
Correlation:0.45138
  |
Correlation:0.45138
  |
Component:299; Jaccard:0.095898
 |
Component:037; Jaccard:0.11528
 |
Component:037; Jaccard:0.13988
 |
Component:037; Jaccard:0.16652
 |
Component:040; Jaccard:0.19308
 |
Component:234; Jaccard:0.21063
 |
Component:234; Jaccard:0.22162
 |
Correlation:0.24028
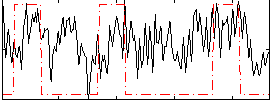 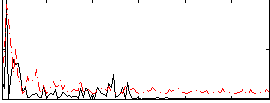 |
Correlation:0.38412
  |
Correlation:0.38412
  |
Correlation:0.38412
  |
Correlation:0.27494
  |
Correlation:0.23244
  |
Correlation:0.23244
  |
Component:037; Jaccard:0.095457
 |
Component:299; Jaccard:0.11072
 |
Component:040; Jaccard:0.13547
 |
Component:040; Jaccard:0.16434
 |
Component:257; Jaccard:0.1923
 |
Component:257; Jaccard:0.20859
 |
Component:257; Jaccard:0.21727
 |
Correlation:0.38412
  |
Correlation:0.24028
  |
Correlation:0.27494
  |
Correlation:0.27494
  |
Correlation:0.25634
  |
Correlation:0.25634
  |
Correlation:0.25634
  |
Component:111; Jaccard:0.093971
 |
Component:040; Jaccard:0.11004
 |
Component:299; Jaccard:0.12733
 |
Component:299; Jaccard:0.14274
 |
Component:299; Jaccard:0.15667
 |
Component:299; Jaccard:0.16582
 |
Component:299; Jaccard:0.17036
 |
Correlation:0.00487
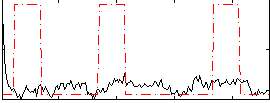 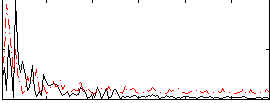 |
Correlation:0.27494
  |
Correlation:0.24028
  |
Correlation:0.24028
  |
Correlation:0.24028
  |
Correlation:0.24028
  |
Correlation:0.24028
  |
Component:242; Jaccard:0.091551
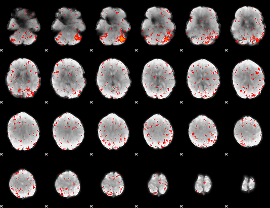 |
Component:242; Jaccard:0.10385
 |
Component:242; Jaccard:0.1162
 |
Component:242; Jaccard:0.12689
 |
Component:242; Jaccard:0.13472
 |
Component:367; Jaccard:0.14111
 |
Component:367; Jaccard:0.1462
 |
Correlation:0.31159
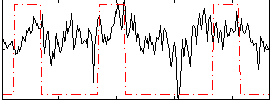 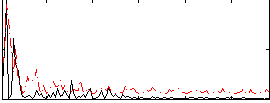 |
Correlation:0.31159
  |
Correlation:0.31159
  |
Correlation:0.31159
  |
Correlation:0.31159
  |
Correlation:0.18269
  |
Correlation:0.18269
  |
Component:400; Jaccard:0.091483
 |
Component:400; Jaccard:0.10083
 |
Component:108; Jaccard:0.1118
 |
Component:108; Jaccard:0.12363
 |
Component:108; Jaccard:0.13319
 |
Component:242; Jaccard:0.13897
 |
Component:108; Jaccard:0.13911
 |
Correlation:0.21376
  |
Correlation:0.21376
  |
Correlation:0.27422
 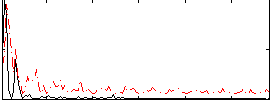 |
Correlation:0.27422
  |
Correlation:0.27422
  |
Correlation:0.31159
  |
Correlation:0.27422
  |
Cope: 03:"MATCH-REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
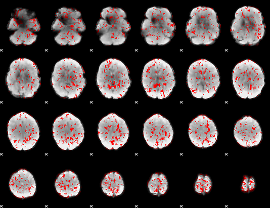 |
GLM binary result thresholded by 1.5
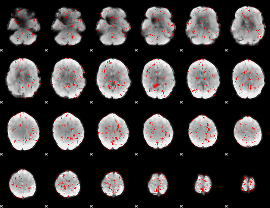 |
GLM binary result thresholded by 2
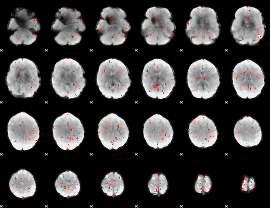 |
GLM binary result thresholded by 2.5
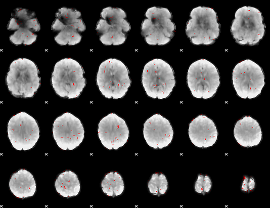 |
GLM binary result thresholded by 3
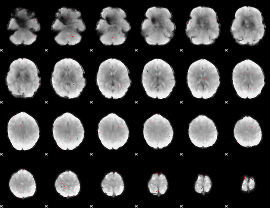 |
GLM binary result thresholded by 3.5
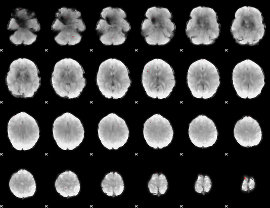 |
GLM binary result thresholded by 4
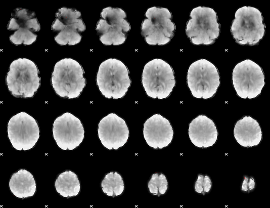 |
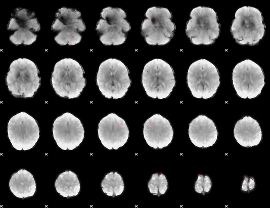
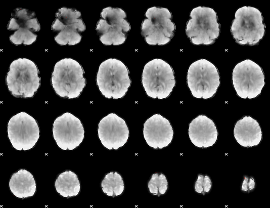
GLM Z-score result thresholded by 1
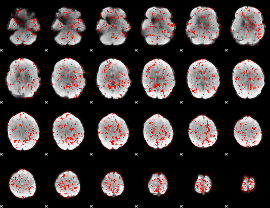 |
GLM Z-score result thresholded by 1.5
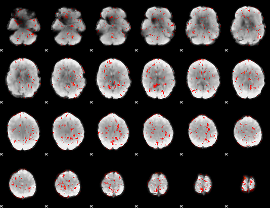 |
GLM Z-score result thresholded by 2
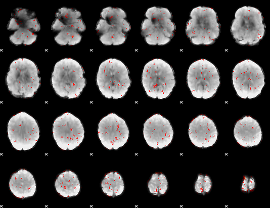 |
GLM Z-score result thresholded by 2.5
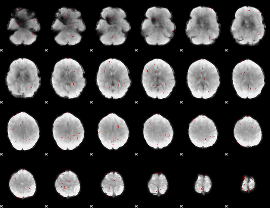 |
GLM Z-score result thresholded by 3
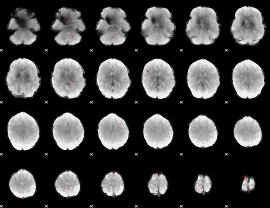 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:360; Jaccard:0.12561
 |
Component:360; Jaccard:0.12749
 |
Component:360; Jaccard:0.10167
 |
Component:086; Jaccard:0.062575
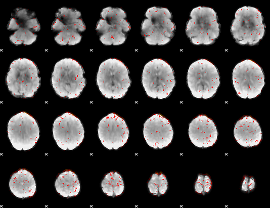 |
Component:086; Jaccard:0.029243
 |
Component:086; Jaccard:0.011236
 |
Component:086; Jaccard:0.003298
 |
Correlation:0.0021514
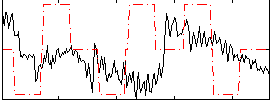 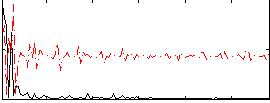 |
Correlation:0.0021514
  |
Correlation:0.0021514
  |
Correlation:0.14303
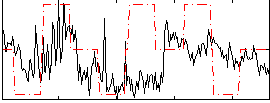 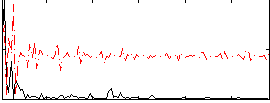 |
Correlation:0.14303
  |
Correlation:0.14303
  |
Correlation:0.14303
  |
Component:086; Jaccard:0.11154
 |
Component:086; Jaccard:0.11873
 |
Component:086; Jaccard:0.10137
 |
Component:360; Jaccard:0.061431
 |
Component:360; Jaccard:0.026941
 |
Component:360; Jaccard:0.008899
 |
Component:169; Jaccard:0.002915
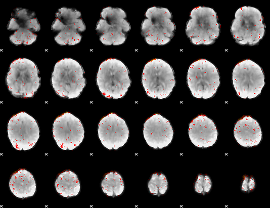 |
Correlation:0.14303
  |
Correlation:0.14303
  |
Correlation:0.14303
  |
Correlation:0.0021514
  |
Correlation:0.0021514
  |
Correlation:0.0021514
  |
Correlation:0.32389
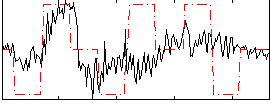 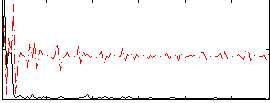 |
Component:023; Jaccard:0.098324
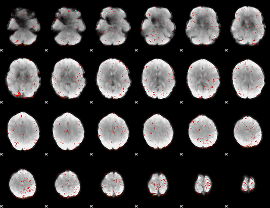 |
Component:169; Jaccard:0.10499
 |
Component:169; Jaccard:0.093758
 |
Component:169; Jaccard:0.060019
 |
Component:169; Jaccard:0.026791
 |
Component:169; Jaccard:0.008626
 |
Component:071; Jaccard:0.00279
 |
Correlation:0.20511
  |
Correlation:0.32389
  |
Correlation:0.32389
  |
Correlation:0.32389
  |
Correlation:0.32389
  |
Correlation:0.32389
  |
Correlation:0.080687
  |
Component:169; Jaccard:0.094895
 |
Component:023; Jaccard:0.099526
 |
Component:185; Jaccard:0.084753
 |
Component:185; Jaccard:0.050863
 |
Component:185; Jaccard:0.020865
 |
Component:071; Jaccard:0.008275
 |
Component:360; Jaccard:0.002302
 |
Correlation:0.32389
  |
Correlation:0.20511
  |
Correlation:0.17284
  |
Correlation:0.17284
  |
Correlation:0.17284
  |
Correlation:0.080687
  |
Correlation:0.0021514
  |
Component:185; Jaccard:0.089274
 |
Component:185; Jaccard:0.098202
 |
Component:023; Jaccard:0.08132
 |
Component:023; Jaccard:0.049008
 |
Component:071; Jaccard:0.019584
 |
Component:185; Jaccard:0.005755
 |
Component:185; Jaccard:0.0019
 |
Correlation:0.17284
  |
Correlation:0.17284
  |
Correlation:0.20511
  |
Correlation:0.20511
  |
Correlation:0.080687
  |
Correlation:0.17284
  |
Correlation:0.17284
  |
Component:365; Jaccard:0.085262
 |
Component:134; Jaccard:0.089116
 |
Component:134; Jaccard:0.076283
 |
Component:134; Jaccard:0.045597
 |
Component:023; Jaccard:0.017954
 |
Component:023; Jaccard:0.005283
 |
Component:023; Jaccard:0.001741
 |
Correlation:-0.14353
  |
Correlation:0.10075
  |
Correlation:0.10075
  |
Correlation:0.10075
  |
Correlation:0.20511
  |
Correlation:0.20511
  |
Correlation:0.20511
  |
Component:134; Jaccard:0.085149
 |
Component:071; Jaccard:0.082157
 |
Component:071; Jaccard:0.06464
 |
Component:071; Jaccard:0.039942
 |
Component:134; Jaccard:0.017927
 |
Component:365; Jaccard:0.004979
 |
Component:365; Jaccard:0.001532
 |
Correlation:0.10075
  |
Correlation:0.080687
  |
Correlation:0.080687
  |
Correlation:0.080687
  |
Correlation:0.10075
  |
Correlation:-0.14353
  |
Correlation:-0.14353
  |
Component:071; Jaccard:0.084024
 |
Component:336; Jaccard:0.078335
 |
Component:365; Jaccard:0.058245
 |
Component:336; Jaccard:0.032421
 |
Component:204; Jaccard:0.015129
 |
Component:175; Jaccard:0.004642
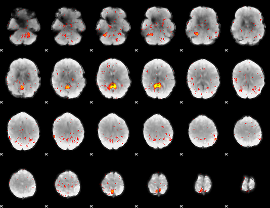 |
Component:175; Jaccard:0.001459
 |
Correlation:0.080687
  |
Correlation:0.077058
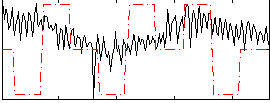 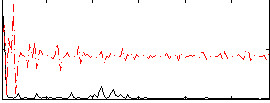 |
Correlation:-0.14353
  |
Correlation:0.077058
  |
Correlation:-0.058407
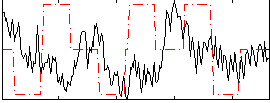  |
Correlation:0.11597
  |
Correlation:0.11597
  |
Component:336; Jaccard:0.080605
 |
Component:365; Jaccard:0.078118
 |
Component:336; Jaccard:0.057501
 |
Component:365; Jaccard:0.03214
 |
Component:175; Jaccard:0.013574
 |
Component:134; Jaccard:0.004556
 |
Component:134; Jaccard:0.001302
 |
Correlation:0.077058
  |
Correlation:-0.14353
  |
Correlation:0.077058
  |
Correlation:-0.14353
  |
Correlation:0.11597
  |
Correlation:0.10075
  |
Correlation:0.10075
  |
Component:002; Jaccard:0.075894
 |
Component:235; Jaccard:0.068359
 |
Component:175; Jaccard:0.051918
 |
Component:204; Jaccard:0.031499
 |
Component:365; Jaccard:0.012737
 |
Component:392; Jaccard:0.004107
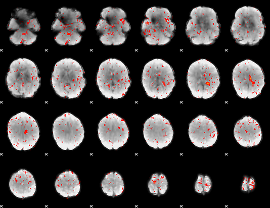 |
Component:281; Jaccard:0.00124
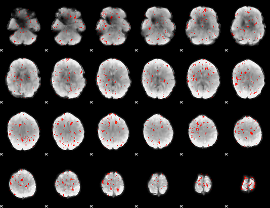 |
Correlation:0.07647
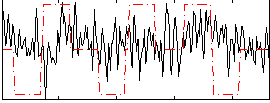 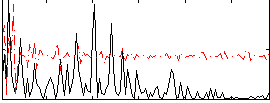 |
Correlation:0.20697
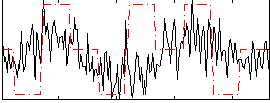 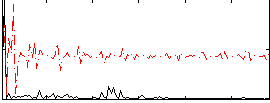 |
Correlation:0.11597
  |
Correlation:-0.058407
  |
Correlation:-0.14353
  |
Correlation:0.085767
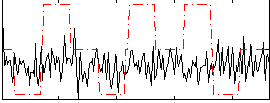 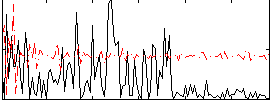 |
Correlation:-0.023306
  |
Cope: 04:"REL-MATCH" BACK
|
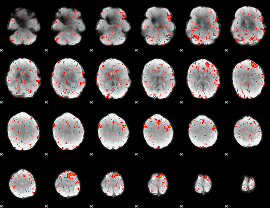
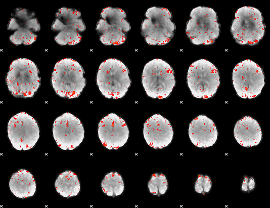
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
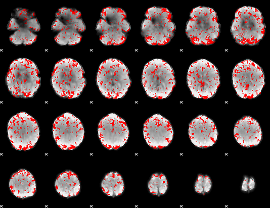 |
GLM binary result thresholded by 2.5
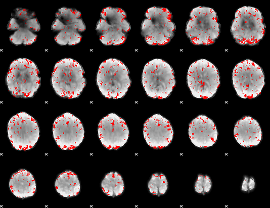 |
GLM binary result thresholded by 3
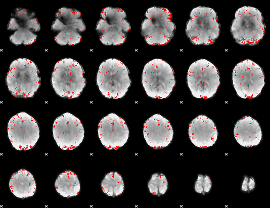 |
GLM binary result thresholded by 3.5
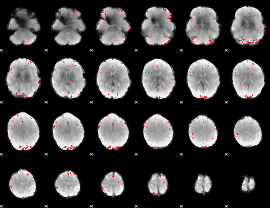 |
GLM binary result thresholded by 4
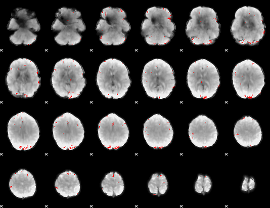 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
 |
GLM Z-score result thresholded by 2
 |
GLM Z-score result thresholded by 2.5
 |
GLM Z-score result thresholded by 3
 |
GLM Z-score result thresholded by 3.5
 |
GLM Z-score result thresholded by 4
 |
Component:010; Jaccard:0.10596
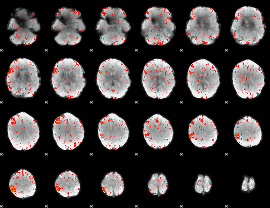 |
Component:108; Jaccard:0.13065
 |
Component:108; Jaccard:0.15731
 |
Component:108; Jaccard:0.17171
 |
Component:108; Jaccard:0.15544
 |
Component:108; Jaccard:0.11333
 |
Component:108; Jaccard:0.064444
 |
Correlation:0.13318
  |
Correlation:0.19378
  |
Correlation:0.19378
  |
Correlation:0.19378
  |
Correlation:0.19378
  |
Correlation:0.19378
  |
Correlation:0.19378
  |
Component:318; Jaccard:0.10556
 |
Component:010; Jaccard:0.12415
 |
Component:387; Jaccard:0.13852
 |
Component:387; Jaccard:0.15168
 |
Component:387; Jaccard:0.13849
 |
Component:387; Jaccard:0.10581
 |
Component:387; Jaccard:0.064275
 |
Correlation:0.10633
  |
Correlation:0.13318
  |
Correlation:0.054805
  |
Correlation:0.054805
  |
Correlation:0.054805
  |
Correlation:0.054805
  |
Correlation:0.054805
  |
Component:108; Jaccard:0.10436
 |
Component:318; Jaccard:0.12105
 |
Component:010; Jaccard:0.13617
 |
Component:010; Jaccard:0.13671
 |
Component:295; Jaccard:0.1161
 |
Component:129; Jaccard:0.088344
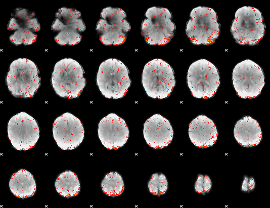 |
Component:129; Jaccard:0.055561
 |
Correlation:0.19378
  |
Correlation:0.10633
  |
Correlation:0.13318
  |
Correlation:0.13318
  |
Correlation:0.22243
  |
Correlation:0.28965
  |
Correlation:0.28965
  |
Component:295; Jaccard:0.097891
 |
Component:387; Jaccard:0.11597
 |
Component:318; Jaccard:0.13049
 |
Component:295; Jaccard:0.12954
 |
Component:010; Jaccard:0.11397
 |
Component:295; Jaccard:0.087827
 |
Component:295; Jaccard:0.055443
 |
Correlation:0.22243
  |
Correlation:0.054805
  |
Correlation:0.10633
  |
Correlation:0.22243
  |
Correlation:0.13318
  |
Correlation:0.22243
  |
Correlation:0.22243
  |
Component:387; Jaccard:0.093535
 |
Component:295; Jaccard:0.11398
 |
Component:295; Jaccard:0.12746
 |
Component:318; Jaccard:0.12675
 |
Component:129; Jaccard:0.11116
 |
Component:037; Jaccard:0.082426
 |
Component:037; Jaccard:0.055098
 |
Correlation:0.054805
  |
Correlation:0.22243
  |
Correlation:0.22243
  |
Correlation:0.10633
  |
Correlation:0.28965
  |
Correlation:0.16615
  |
Correlation:0.16615
  |
Component:129; Jaccard:0.090797
 |
Component:037; Jaccard:0.10549
 |
Component:129; Jaccard:0.1182
 |
Component:391; Jaccard:0.12486
 |
Component:391; Jaccard:0.10988
 |
Component:391; Jaccard:0.078515
 |
Component:391; Jaccard:0.04503
 |
Correlation:0.28965
  |
Correlation:0.16615
  |
Correlation:0.28965
  |
Correlation:0.30745
  |
Correlation:0.30745
  |
Correlation:0.30745
  |
Correlation:0.30745
  |
Component:037; Jaccard:0.0907
 |
Component:129; Jaccard:0.10469
 |
Component:037; Jaccard:0.11741
 |
Component:129; Jaccard:0.12231
 |
Component:037; Jaccard:0.10845
 |
Component:010; Jaccard:0.076177
 |
Component:010; Jaccard:0.043347
 |
Correlation:0.16615
  |
Correlation:0.28965
  |
Correlation:0.16615
  |
Correlation:0.28965
  |
Correlation:0.16615
  |
Correlation:0.13318
  |
Correlation:0.13318
  |
Component:354; Jaccard:0.089225
 |
Component:391; Jaccard:0.099928
 |
Component:391; Jaccard:0.11692
 |
Component:037; Jaccard:0.12026
 |
Component:318; Jaccard:0.10135
 |
Component:318; Jaccard:0.069314
 |
Component:048; Jaccard:0.040583
 |
Correlation:0.2194
  |
Correlation:0.30745
  |
Correlation:0.30745
  |
Correlation:0.16615
  |
Correlation:0.10633
  |
Correlation:0.10633
  |
Correlation:0.038412
  |
Component:257; Jaccard:0.088899
 |
Component:257; Jaccard:0.096607
 |
Component:257; Jaccard:0.098907
 |
Component:257; Jaccard:0.09099
 |
Component:181; Jaccard:0.078858
 |
Component:181; Jaccard:0.063779
 |
Component:181; Jaccard:0.040412
 |
Correlation:0.088908
  |
Correlation:0.088908
  |
Correlation:0.088908
  |
Correlation:0.088908
  |
Correlation:0.010831
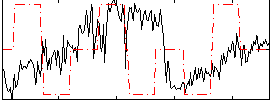 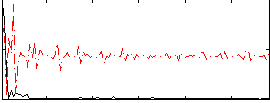 |
Correlation:0.010831
  |
Correlation:0.010831
  |
Component:258; Jaccard:0.083504
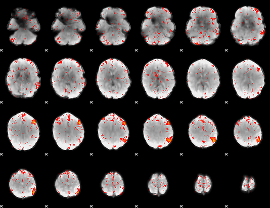 |
Component:354; Jaccard:0.094253
 |
Component:354; Jaccard:0.09312
 |
Component:048; Jaccard:0.089987
 |
Component:048; Jaccard:0.078547
 |
Component:048; Jaccard:0.060055
 |
Component:318; Jaccard:0.039687
 |
Correlation:0.21451
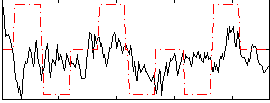  |
Correlation:0.2194
  |
Correlation:0.2194
  |
Correlation:0.038412
  |
Correlation:0.038412
  |
Correlation:0.038412
  |
Correlation:0.10633
  |
Cope: 05:"neg_MATCH" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
 |
GLM binary result thresholded by 2.5
 |
GLM binary result thresholded by 3
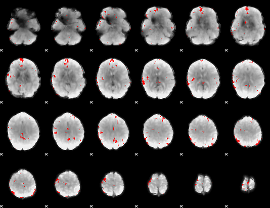 |
GLM binary result thresholded by 3.5
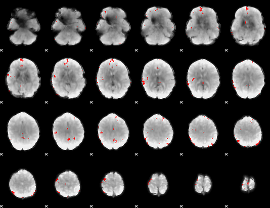 |
GLM binary result thresholded by 4
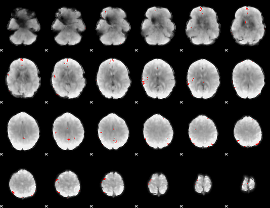 |
GLM Z-score result thresholded by 1
 |
GLM Z-score result thresholded by 1.5
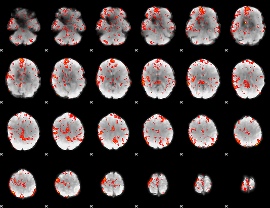 |
GLM Z-score result thresholded by 2
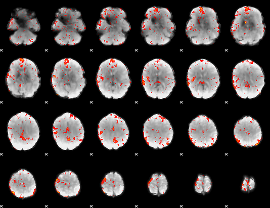 |
GLM Z-score result thresholded by 2.5
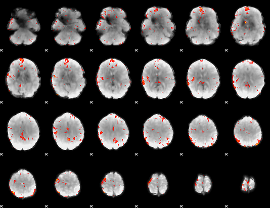 |
GLM Z-score result thresholded by 3
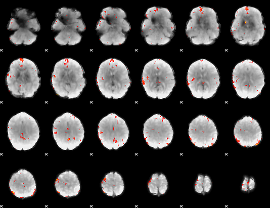 |
GLM Z-score result thresholded by 3.5
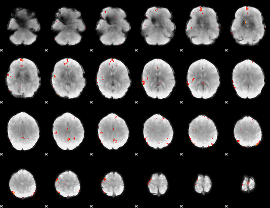 |
GLM Z-score result thresholded by 4
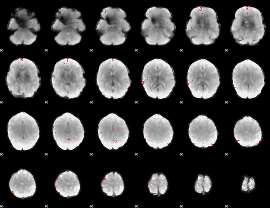 |
Component:107; Jaccard:0.14626
 |
Component:107; Jaccard:0.16618
 |
Component:107; Jaccard:0.17206
 |
Component:107; Jaccard:0.15248
 |
Component:107; Jaccard:0.11692
 |
Component:107; Jaccard:0.074218
 |
Component:107; Jaccard:0.041129
 |
Correlation:0.21361
 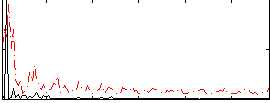 |
Correlation:0.21361
  |
Correlation:0.21361
  |
Correlation:0.21361
  |
Correlation:0.21361
  |
Correlation:0.21361
  |
Correlation:0.21361
  |
Component:344; Jaccard:0.12372
 |
Component:344; Jaccard:0.13986
 |
Component:344; Jaccard:0.14396
 |
Component:344; Jaccard:0.12739
 |
Component:344; Jaccard:0.094194
 |
Component:344; Jaccard:0.062625
 |
Component:344; Jaccard:0.036516
 |
Correlation:0.27211
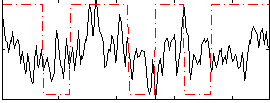 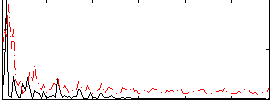 |
Correlation:0.27211
  |
Correlation:0.27211
  |
Correlation:0.27211
  |
Correlation:0.27211
  |
Correlation:0.27211
  |
Correlation:0.27211
  |
Component:153; Jaccard:0.11429
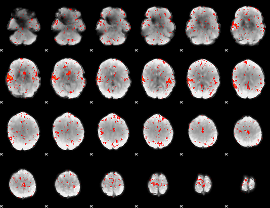 |
Component:105; Jaccard:0.11939
 |
Component:105; Jaccard:0.12059
 |
Component:105; Jaccard:0.10142
 |
Component:135; Jaccard:0.074866
 |
Component:135; Jaccard:0.051906
 |
Component:135; Jaccard:0.030705
 |
Correlation:0.22872
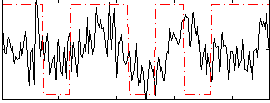 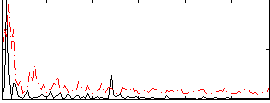 |
Correlation:0.25682
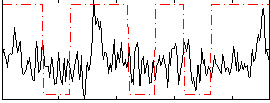  |
Correlation:0.25682
  |
Correlation:0.25682
  |
Correlation:0.22284
  |
Correlation:0.22284
  |
Correlation:0.22284
  |
Component:105; Jaccard:0.10916
 |
Component:153; Jaccard:0.11888
 |
Component:153; Jaccard:0.11104
 |
Component:135; Jaccard:0.093676
 |
Component:105; Jaccard:0.073617
 |
Component:105; Jaccard:0.047521
 |
Component:156; Jaccard:0.030123
 |
Correlation:0.25682
  |
Correlation:0.22872
  |
Correlation:0.22872
  |
Correlation:0.22284
  |
Correlation:0.25682
  |
Correlation:0.25682
  |
Correlation:0.20048
  |
Component:082; Jaccard:0.094693
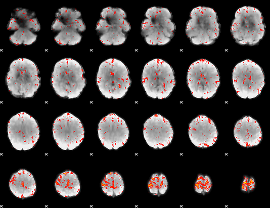 |
Component:135; Jaccard:0.10338
 |
Component:135; Jaccard:0.10421
 |
Component:153; Jaccard:0.086614
 |
Component:219; Jaccard:0.065202
 |
Component:156; Jaccard:0.04365
 |
Component:105; Jaccard:0.026456
 |
Correlation:0.14403
  |
Correlation:0.22284
  |
Correlation:0.22284
  |
Correlation:0.22872
  |
Correlation:0.34003
  |
Correlation:0.20048
  |
Correlation:0.25682
  |
Component:279; Jaccard:0.094177
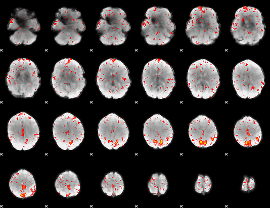 |
Component:219; Jaccard:0.097811
 |
Component:219; Jaccard:0.098347
 |
Component:219; Jaccard:0.086312
 |
Component:153; Jaccard:0.058194
 |
Component:219; Jaccard:0.040144
 |
Component:113; Jaccard:0.024754
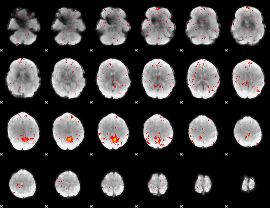 |
Correlation:0.099766
 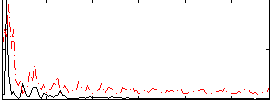 |
Correlation:0.34003
  |
Correlation:0.34003
  |
Correlation:0.34003
  |
Correlation:0.22872
  |
Correlation:0.34003
  |
Correlation:0.15267
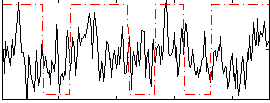 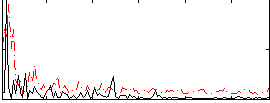 |
Component:135; Jaccard:0.093301
 |
Component:279; Jaccard:0.093584
 |
Component:109; Jaccard:0.090889
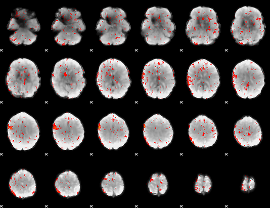 |
Component:109; Jaccard:0.077386
 |
Component:156; Jaccard:0.057934
 |
Component:204; Jaccard:0.038666
 |
Component:109; Jaccard:0.022844
 |
Correlation:0.22284
  |
Correlation:0.099766
  |
Correlation:0.1491
  |
Correlation:0.1491
  |
Correlation:0.20048
  |
Correlation:0.24832
  |
Correlation:0.1491
  |
Component:354; Jaccard:0.092177
 |
Component:354; Jaccard:0.090432
 |
Component:279; Jaccard:0.084542
 |
Component:204; Jaccard:0.075589
 |
Component:109; Jaccard:0.057502
 |
Component:109; Jaccard:0.037341
 |
Component:204; Jaccard:0.022535
 |
Correlation:0.1462
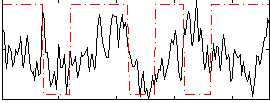 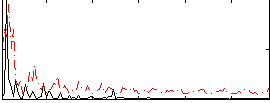 |
Correlation:0.1462
  |
Correlation:0.099766
  |
Correlation:0.24832
  |
Correlation:0.1491
  |
Correlation:0.1491
  |
Correlation:0.24832
  |
Component:219; Jaccard:0.090147
 |
Component:082; Jaccard:0.090378
 |
Component:204; Jaccard:0.08216
 |
Component:376; Jaccard:0.073898
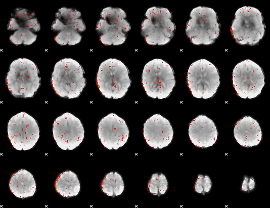 |
Component:204; Jaccard:0.056484
 |
Component:113; Jaccard:0.037098
 |
Component:219; Jaccard:0.022453
 |
Correlation:0.34003
  |
Correlation:0.14403
  |
Correlation:0.24832
  |
Correlation:0.22109
 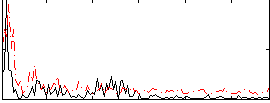 |
Correlation:0.24832
  |
Correlation:0.15267
  |
Correlation:0.34003
  |
Component:192; Jaccard:0.087369
 |
Component:109; Jaccard:0.0896
 |
Component:156; Jaccard:0.079121
 |
Component:156; Jaccard:0.070412
 |
Component:376; Jaccard:0.055005
 |
Component:153; Jaccard:0.036721
 |
Component:376; Jaccard:0.020496
 |
Correlation:0.25208
  |
Correlation:0.1491
  |
Correlation:0.20048
  |
Correlation:0.20048
  |
Correlation:0.22109
  |
Correlation:0.22872
  |
Correlation:0.22109
  |
Cope: 06:"neg_REL" BACK
|
GLM result group-wise
 |
GLM binary result thresholded by 1
 |
GLM binary result thresholded by 1.5
 |
GLM binary result thresholded by 2
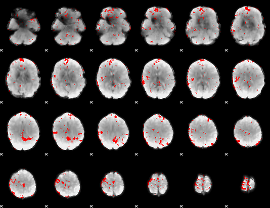 |
GLM binary result thresholded by 2.5
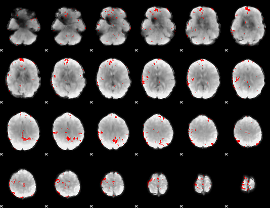 |
GLM binary result thresholded by 3
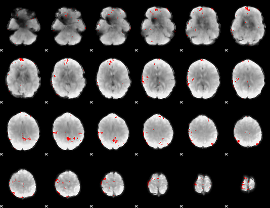 |
GLM binary result thresholded by 3.5
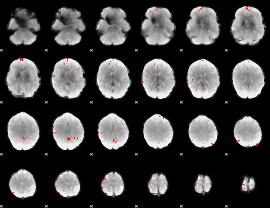 |
GLM binary result thresholded by 4
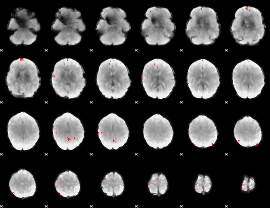 |
GLM Z-score result thresholded by 1
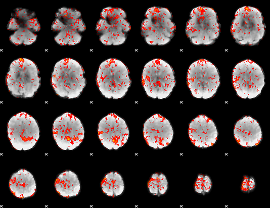 |
GLM Z-score result thresholded by 1.5
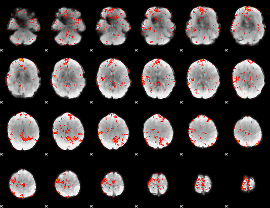 |
GLM Z-score result thresholded by 2
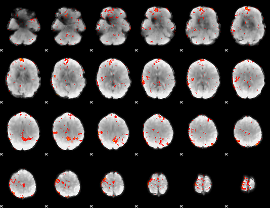 |
GLM Z-score result thresholded by 2.5
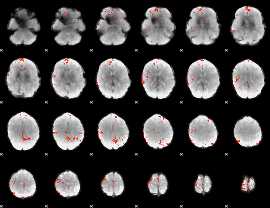 |
GLM Z-score result thresholded by 3
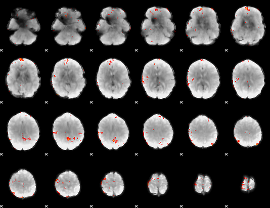 |
GLM Z-score result thresholded by 3.5
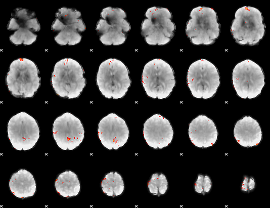 |
GLM Z-score result thresholded by 4
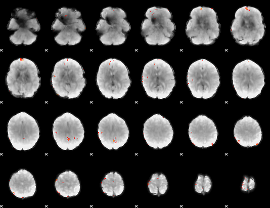 |
Component:107; Jaccard:0.13639
 |
Component:107; Jaccard:0.14537
 |
Component:135; Jaccard:0.14939
 |
Component:135; Jaccard:0.13417
 |
Component:135; Jaccard:0.10408
 |
Component:204; Jaccard:0.074729
 |
Component:204; Jaccard:0.048428
 |
Correlation:-0.026045
  |
Correlation:-0.026045
  |
Correlation:0.14861
  |
Correlation:0.14861
  |
Correlation:0.14861
  |
Correlation:0.14977
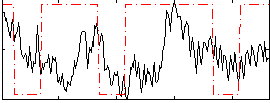 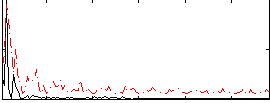 |
Correlation:0.14977
  |
Component:135; Jaccard:0.12361
 |
Component:135; Jaccard:0.14073
 |
Component:107; Jaccard:0.14037
 |
Component:204; Jaccard:0.12701
 |
Component:204; Jaccard:0.10396
 |
Component:135; Jaccard:0.072217
 |
Component:135; Jaccard:0.042521
 |
Correlation:0.14861
  |
Correlation:0.14861
  |
Correlation:-0.026045
  |
Correlation:0.14977
  |
Correlation:0.14977
  |
Correlation:0.14861
  |
Correlation:0.14861
  |
Component:082; Jaccard:0.11085
 |
Component:204; Jaccard:0.12682
 |
Component:204; Jaccard:0.13494
 |
Component:107; Jaccard:0.12421
 |
Component:107; Jaccard:0.088385
 |
Component:107; Jaccard:0.059975
 |
Component:107; Jaccard:0.037328
 |
Correlation:0.1752
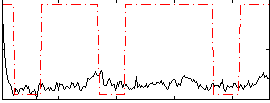 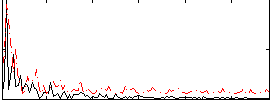 |
Correlation:0.14977
  |
Correlation:0.14977
  |
Correlation:-0.026045
  |
Correlation:-0.026045
  |
Correlation:-0.026045
  |
Correlation:-0.026045
  |
Component:204; Jaccard:0.10781
 |
Component:365; Jaccard:0.10934
 |
Component:219; Jaccard:0.10319
 |
Component:219; Jaccard:0.090409
 |
Component:113; Jaccard:0.068424
 |
Component:113; Jaccard:0.04778
 |
Component:219; Jaccard:0.030576
 |
Correlation:0.14977
  |
Correlation:0.042614
  |
Correlation:-0.11743
  |
Correlation:-0.11743
  |
Correlation:0.23469
  |
Correlation:0.23469
  |
Correlation:-0.11743
  |
Component:365; Jaccard:0.10728
 |
Component:082; Jaccard:0.10467
 |
Component:365; Jaccard:0.099681
 |
Component:344; Jaccard:0.084183
 |
Component:219; Jaccard:0.068036
 |
Component:219; Jaccard:0.046523
 |
Component:113; Jaccard:0.028257
 |
Correlation:0.042614
  |
Correlation:0.1752
  |
Correlation:0.042614
  |
Correlation:0.065765
  |
Correlation:-0.11743
  |
Correlation:-0.11743
  |
Correlation:0.23469
  |
Component:344; Jaccard:0.10219
 |
Component:344; Jaccard:0.10425
 |
Component:344; Jaccard:0.09778
 |
Component:113; Jaccard:0.082835
 |
Component:344; Jaccard:0.060411
 |
Component:344; Jaccard:0.037676
 |
Component:365; Jaccard:0.021331
 |
Correlation:0.065765
  |
Correlation:0.065765
  |
Correlation:0.065765
  |
Correlation:0.23469
  |
Correlation:0.065765
  |
Correlation:0.065765
  |
Correlation:0.042614
  |
Component:360; Jaccard:0.099858
 |
Component:219; Jaccard:0.10409
 |
Component:360; Jaccard:0.094212
 |
Component:365; Jaccard:0.080003
 |
Component:365; Jaccard:0.057093
 |
Component:365; Jaccard:0.035309
 |
Component:344; Jaccard:0.021284
 |
Correlation:0.086288
  |
Correlation:-0.11743
  |
Correlation:0.086288
  |
Correlation:0.042614
  |
Correlation:0.042614
  |
Correlation:0.042614
  |
Correlation:0.065765
  |
Component:219; Jaccard:0.09832
 |
Component:360; Jaccard:0.10153
 |
Component:082; Jaccard:0.089437
 |
Component:360; Jaccard:0.076368
 |
Component:360; Jaccard:0.054401
 |
Component:360; Jaccard:0.034476
 |
Component:249; Jaccard:0.021162
 |
Correlation:-0.11743
  |
Correlation:0.086288
  |
Correlation:0.1752
  |
Correlation:0.086288
  |
Correlation:0.086288
  |
Correlation:0.086288
  |
Correlation:0.11322
  |
Component:106; Jaccard:0.09286
 |
Component:105; Jaccard:0.092352
 |
Component:113; Jaccard:0.086518
 |
Component:105; Jaccard:0.069192
 |
Component:105; Jaccard:0.051156
 |
Component:249; Jaccard:0.032712
 |
Component:360; Jaccard:0.019156
 |
Correlation:-0.050121
  |
Correlation:-0.056497
  |
Correlation:0.23469
  |
Correlation:-0.056497
  |
Correlation:-0.056497
  |
Correlation:0.11322
  |
Correlation:0.086288
  |
Component:105; Jaccard:0.092579
 |
Component:106; Jaccard:0.089591
 |
Component:105; Jaccard:0.084863
 |
Component:082; Jaccard:0.067592
 |
Component:249; Jaccard:0.048135
 |
Component:105; Jaccard:0.031524
 |
Component:352; Jaccard:0.018739
 |
Correlation:-0.056497
  |
Correlation:-0.050121
  |
Correlation:-0.056497
  |
Correlation:0.1752
  |
Correlation:0.11322
  |
Correlation:-0.056497
  |
Correlation:-0.091897
  |





































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































































